Journal of
eISSN: 2377-4312


Review Article Volume 5 Issue 5
Department of Animal, Aquaculture and Range Sciences (DAARS), Sokoine University of Agriculture, Tanzania
Correspondence: G Msalya, Department of Animal Science and Production, Sokoine University of Agriculture, P.O. Box 3004, Morogoro, Tanzania, Tel 25 5232 6035 1114, Fax 2552 3260 4562
Received: April 10, 2017 | Published: July 21, 2017
Citation: Msalya G, Lutatenekwa D, Chenyambuga SW. Possibilities of utilizing biotechnology to improve animal and animal feeds productivity in Tanzania – review of past efforts and available opportunities. J Dairy Vet Anim Res. 2017;5(5):161-167. DOI: 10.15406/jdvar.2017.05.00155
Tanzania ranks third in Africa in numbers of livestock and has a rich biodiversity of 25.8milion heads of cattle, about 25million heads of small ruminants (sheep and goats) and a large number of other livestock species. The majority of livestock species are indigenous animals with low productive potential. In addition, the farming systems are still to a large extent traditional utilizing local feeds which have very low quality. Due to these reasons the contribution of livestock sector to the gross domestic product has been very low estimated between 4.7 and 5%. There have been attempts to improve productivity from livestock and livestock feeds in the past through different research and development projects. However most of these projects were not sustainable and most of them did not bear good fruits while some were abandoned. Biotechnology offers options for improvement of the livestock sector by manipulating both livestock species and management systems such as the environment and feeds. However, there is a very low rate of investment in biotechnology especially in livestock and feeds production. In this paper we review, the efforts intended at improvement of livestock genetics and animal nutrition and we discuss the options and opportunities for improvement of the livestock sector using modern technologies. Examples of these efforts include the breeding of Mpwapwa cattle, Malya blended goats and improvements of various breeds of other livestock species and poultry. We showcase these possibilities using three studies conducted recently in Tanzania, including: (1) characterization of the indigenous stock of cattle to develop breeding plans, (2) identification of pathogens in raw milk in the informal markets of Tanzania, and (3) expression of the prion protein gene for resistance to bovine spongiform encephalopathy (BSE).
Keywords: tanzania shorthorn zebu, small east african goats, breeding, productivity, biotechnology
AI, artificial insemination; ANFs, anti-nutritional factors; BHP, black head persian (sheep); BSE, bovine spongiform encephalopathy; EAZ, east african zebu; ET, embryo transfer; FAO, food and agriculture organization; LN, liquid nitrogen; MAS, marker assisted selection; MARI, mikocheni agricultural research institute; MOET, multiple ovulation and embryo transfer; MPTs, multipurpose trees; NAIC, national artificial insemination centre; NARCO, national ranching company; PCR, polymerized chain reaction; PRNP, prion protein gene; SEA, small east african (goats); SFC, sunflower cake; SNP, single nucleotide polymorphism; SUA, sokoine university of agriculture; TBDs, tick-borne diseases; TVLA, tanzania veterinary laboratory agency; TZS, tanzania shorthorn zebu (cattle); UNCED, united nations conference on environment and development
Recently annual per capita intake of animal protein in Tanzania was estimated to be 15kg of meat, 47litres of milk and 106 eggs. These figures are lower compared to the rates recommended by United Nation’s Food and Agriculture Organization (FAO) of 50kg of meat, 200litres of milk and 300 eggs per person per year.1 As a result, Tanzania was listed among top 10 countries in the world and third in Africa in number of undernourished children.2 Problem of food insecurity including animal protein deficiency in particular among children under five years of age and pregnant women is expected to widen further ahead 2050.2 At the same time, the country is ranked third in Africa in numbers of livestock with estimated 25.8milion heads of cattle, 16.7million goats, 8.7million sheep, 2.4million pigs and about 70million chickens. Furthermore, production of animal products such as meat, milk and eggs has increased significantly in recent years. For example, meat production has increased by over 6.2percent to 597,757 tonnes in 2015.1 Milk production increased by 130% over the last decade to about 2.25billion litres in 20153 and over 4billion eggs were estimated in 2015.1 However, it is generally agreed that the increases have not met the greater demand for these products evidenced by the low consumption rates and hiking prices in local markets in the country. Also, the increases are due to increase in number of animals but not productivity per animal.3 A larger percent of animals raised in Tanzania belong to the native populations which have been characterized to be low producers of meat, milk and eggs due to poor genetic potential.4‒6 For example, Tanzania Shorthorn Zebu (TSZ) animals which form 95% of all cattle and produce over 70% and 90% of milk and meat respectively are characterized by slow growth rates, low mature weight, long calving intervals, as well as low meat and milk yield.1 Of the 16.7million goats about 98% belongs to the native Small East African (SEA) breed estimated to have low growth rates between 5 and 7g/day, small mature size ranging from 20 to 25kg and low carcass weights about 6 to 13kg.6,7 Likewise productivity levels and reproductive efficiency are also far below optimum ranges in other species. Therefore, strategies for improvements are needed in order to improve performance and productivity of the local genetic resources.
Low production is also contributed by poor and unfavorable production environments including lack of good quality and optimum amount of feeds leading to poor nutrition as well as the occurrence of diseases and parasites. For example, rangelands are the major source of feeds for ruminants in the country. However, degradation of rangelands due to severe stocking rates, weather uncertainty, and impacts of climate change have resulted to limited quantity and quality of feeds for these animals year after year.8,9 This situation causes seasonal variations in feeds and results to growth fluctuations of animals and prolonged time to reach market weights, consequently leading to production of poor quality meat. In addition, ruminants are unable to meet their maintenance and production requirements, especially for protein and therefore there have been low growth rates, low conception rates, high neonatal losses and high incidences of diseases recorded in different farming systems.1 Regarding diseases, tick-borne diseases (TBDs) were reported to affect about 80% of the total cattle herds with direct economic losses of about 248million US$ and close to one million deaths per year until recently.10,11 These and other factors negatively affect the animal production sector and it is therefore difficult to effectively improve animal production based on low quality nutrition of native unimproved feeds and conventional methods of disease control.
Biotechnology offers ample opportunity of improving the genetics and productivity of livestock species through direct selection using genomic sequencing and analyses, reducing incidences of diseases by direct diagnosis, and increasing tolerance to unfavourable environment and climatic conditions. Furthermore, it has the potential to improve animal feeds or digestive systems of livestock species. However, there is a very low rate of investment in biotechnology in livestock sector in general. In this paper we review, the development of biotechnology in livestock production sector of Tanzania (past efforts to improve livestock and feeds production or examples of what has been done, as well as the present opportunities or what can be further achieved). We also showcase the use of biotechnology in characterization of the indigenous stock of cattle using single nucleotide polymorphism (SNP) markers and their potential in develop breeding programmes, identification of pathogens in raw milk in the informal markets of Tanzania, and expression of the prion protein gene for resistance to bovine spongiform encephalopathy (BSE).
Past attempts to improve productivity in the livestock sector in Tanzania (genetics and feeds)
Cattle selection and breeding: Genetic improvement programmes started as early as mid-1920s when local cattle were phenotypically characterized and selection within populations was implemented. One of the earliest breeding objectives was improvement of milk production through selective breeding within the indigenous (Tanganyika Zebu) cattle which continued until 1940s,12 leading to production of animals which were adapted to the tropical environment. Selection was based on culling standards and essentially milk yield per lactation gradually increased from 460kg in 1940s to 920kg (approximately doubled) in mid 1950s and it was apparent that selective breeding within local strains can produce the type of animals needed.13 However, encouraging progress, similar programmes in Kenya and Uganda showed increase of milk production among East African Zebu (EAZ) from 530 to maximum of 950kg per lactation leading to conclusions that these animals (the EAZ including TSZ) were lower producers of milk.14‒17 Because of the low genetic capability of the TSZ, crossbreeding with high producing taurines started in 1930s. The TSZ females were mated to Friesian and Ayrshire with the intention of grading up the TSZ for milk production. The first experiments were conducted in Dar es Salaam (Temeke Dairy Farm) and in Dodoma at Mpwapwa research centre involving crossbreeding of TSZ with Ayrshire and progeny interbreeding in 1932. However, disappointing lactation performance levels of high-grade cattle were reported.18,19 A few years later Boran breed was imported into the country and was selected and bred for milk production in Tanga region.20 From 1970s to 1990s, importations of exotic livestock continued to be driven by high production motives and were not linked to meaningful sustainable breeding programmes. Consequently, the growth of exotic and crossbred herds has been limited, thus necessitating continuous importation of replacement exotic breeds. The current population of exotic dairy cattle is composed of crossbreds of Friesian, Ayrshire, and Jerseys.
Earlier reports selection for growth and carcass characteristics showed that TSZ cattle could reach live weight of 425kg at maturity with mean weights of 370kg (males) and 275kg (females) at three years for well-fed cattle.12 Also several breeds including Africander, Krishna Valley, Shorthorn, Sahiwal Red Sindhi, and Devon were mated with the TSZ to improve meat production at the Veterinary department in Dar es Salaam and at the research stations in Dodoma.18 Only Shorthorn x TSZ crossbreds gave fairly favourable results in terms of conformation and fitness than the other crossbreds. Frenche et al.19 observed that upgrading beyond the F1 level did not achieve the higher production as expected, in fact the three-quarter bred and higher grades were less economical than the better TSZ.19 More high producing breeds including Hereford, Charolais, Simmental, Angus, and Chianina were imported and used for crossbreeding with TSZ and Boran cows in 1960s and 1970s.21 The animals were raised in the ranches and earlier studies showed marginal superiority of the crosses in comparison to TSZ in traits such as slaughter weight and cold carcass weight, but not in dressing percentage. To date only a few crosses of these breeds with TSZ or Borans are raised in National Ranching Company (NARCO) ranches. Consequently Boran crossbreds appeared to be more advantageous and greater number of them is found in NARCO ranches.22,23
Probably the most remarkable example of breeding was the formation of a synthetic dual purpose Mpwapwa breed in 1958 with a goal of obtaining yield of 2300kg milk in a standard lactation of 305days and carcass yield of about 230kg from steers of 4years without much dependence on concentrate feeds.24,25 The composition of Mpwapwa cattle was 32% Red Sindhi, 30% Sahiwal, 19% TSZ, 11% Boran and 8 to 10% B. Taurus (Ayrshire, Friesian, Jersey and Guernsey). However, the performance of Mpwapwa animals has been found to be below the expected and these animals were not spread in the traditional sector because they easily succumbed to TBDs.23
Small ruminants breeding: Regarding goats, experiments were undertaken to improve meat production traits by crossbreeding Boer goats and Kamorai with SEA goats. Similarly, crossbreeding was performed between exotic goat breeds such as Toggenburg, British Alpine, Anglo–Nubian and SEA goats to improve milk production under Tanzanian conditions. One important experiment involving three breeds; SEA, Boer and Kamorai was conducted at Mpwapwa and Malya in 1950s leading to formation of a three-way dual purpose goat breed, known as the ‘Malya Blended’ goat. Also, the performance is not very encouraging and to date Malya blended goats are extinct whereas purebred and crossbred goats, notably Toggenburg, Anglo-Nubian, Saanen, Norwegian and Boer are found in small numbers (less than 2% of all goats) and their distribution is limited to high potential highland areas.1,26 Concerning sheep, very little work has been done. In the 1960s Black Head Persian (BHP), Corridale and Merino breeds were introduced in the country. The BHP was intended for improved mutton production in commercial ranches, while the other two breeds were intended for wool production. The spread of BHP sheep over the years has not been very effective and it has mainly remained in few commercial ranches. Farmers near the ranches have, however, used BHP rams to breed with indigenous sheep. The wool sheep were limited to the cold Southern Highlands of Tanzania, but the breeding programme for these sheep was not sustained, as a result there is currently no trace of them in the country. The local uncharacterized sheep can be seen in large numbers in the country.
Pigs and chicken breeding: There is no evidence of existence of indigenous pigs in Tanzania. Commercial pig production is based on the Large White, Large Black, Saddleback, Hampshire and the Landrace breeds which were imported into the country to improve pork production. However, there have not been major organized breeding programmes for pigs in the country. This has resulted in haphazard crossing with little control or guidance from government or breed associations. With regard to chickens the most popular exotic chickens have been the Rhode Island Red, White Leghorn, Light Sussex, Black Australop and various layer and broiler hybrids. Crossbreeding trials have been carried out between indigenous chickens and exotic breeds for meat and egg production. As for the case with pigs, there have not been any sustained breeding programmes for poultry. There is currently a shift in preference from exotic to local chicken product consumption. This calls for efforts towards characterization, improvement and conservation of local chickens.
Improvement in animal nutrition: Strategies to improve nutrition have done mainly for ruminants. Among the strategies include treatment of low quality roughage with urea and the use of conventional supplements.27,28 Tremendous improvement in growth rate (from 3 to 49g/day) was documented in growing dairy goats fed urea treated rice straw supplemented with fishmeal and rice polishing.28 Also feeding of alkaline treated rice straw to sheep (2years) resulted in increased growth rate of 85g per day.29 However, conventional protein supplements such as oil cakes, fishmeal, cereals and their by-products are limited in supply or too expensive to be afforded by livestock keepers in rural areas. Supplementary strategies using cheap and locally available feeds in rural areas are therefore needed. For this reason, leguminous tree leaves and pods involving the use of multipurpose trees (MPTs) such as Leucaena leucocephala and Moringa oleifera30,31 has been the most appropriate alternative protein and energy sources to conventional supplements. Ndemanisho et al. recommended that Leucaena leucocephala leaf meal could substitute two thirds of cotton seed cake to weaners’ goats without reduction in performance.30Also Sarwatt et al. suggested that Moringa oleifera leaf meal could be used to substitute sunflower cake (SFC).31 However, the leguminous trees need to be established as some of them are not indigenous to Tanzania. In other studies, indigenous trees and shrubs leaf meals were used to replace conventional supplements like SFC and were shown to have high growth rates in goats (21g/day) compared to 7g/day for goats fed SFC.32 Indigenous plants have also been shown to possess medicinal effects in the animals.33
Overview of biotechnological development and opportunities for Tanzania: Biotechnology has been a subject of great public interest since late 1980s and was recognized by United Nations Conference on Environment and Development (UNCED) in 1992 that it can make a significant contribution in agricultural production improvement. In Tanzania biotechnology offers the potential for increasing and improving food production capacity and promoting sustainability. Furthermore, biotechnology can be employed in conservation of natural resources including indigenous animal genetic resources. Most importantly, biotechnology can significantly improve animal production, both yield and quality of products and therefore can become one of the strategies for improving animal protein production. This can be achieved through development of animals with enhanced productivity or ones which are tolerant to environmental stresses, such as diseases and pests, as well as drought and high temperatures. The techniques can also be used to improve feed conversion efficiency or improve digestibility of low quality roughages and other animal feed resources. However, application of biotechnology in Tanzania is still at infancy and is only limited to few public institutions engaged in biotechnology research including Sokoine University of Agriculture (SUA), Mikocheni Agricultural Research Institute (MARI), and Tanzania Veterinary Laboratory Agency (TVLA). In these institutions, the most used techniques are limited to tissue culture and micropropagation, biofertilizers and biopesticides, molecular diagnostics for plant and animal pathogens, DNA marker technology and recombinant vaccines. Several biotechnological tools can be successfully applied in Tanzania to improve animal and feeds production.
Artificial insemination: Through artificial insemination (AI) it is possible to collect, process and preserve semen from selected bulls for use to proven progenies. Because of this one bull can enable a large number of inseminations in different times and locations at the same time or at different times.34 At the present time, semen sexing has made it possible to decide sex of the progenies.35 In Tanzania, the National artificial insemination centre (NAIC) was established in 1958 and is the only centre with a mandate to produce, control quality, preserve, and distribute semen. It also provides all AI equipments, train inseminators, collect and keep AI records all over the country. According to NAIC reports semen is collected from Ayrshire, Friesian, Jersey, Simmental, Boran, Sahiwal and Mpwapwa bulls and processed at the centre and until 2014, distribution of 236,335 doses of semen and 47,827.95 litres of liquid nitrogen (LN) to various organizations or farmers were recorded. All programmes at NAIC were designed for the purpose of upgrading the native (indigenous) livestock of Tanzania. However, this technology is only implemented by medium and large scale dairy and beef production farms in the country. It is either rare (limited) or not implemented among pastoralists communities and probably not practiced in other stock such as sheep, goats, pigs and chicken. If promoted and implemented throughout the country positive changes in the livestock sector will be realized.
Embryo transfer: Embryo transfer (ET) can facilitate improvement of genetic potential in animals. According to Taylor et al.36 ET technology can be followed by cloning and embryo sexing to increase reproduction efficiency.36 Also, through multiple ovulation and embryo transfer (MOET) it has been possible to perform in vitro culture of embryos and enable freezing of embryos from rare populations or endangered breeds or species. Application of these technologies can increase genetic gains37 and production of progenies.38,39 In Tanzania as in other developing countries ET is generally not widely used due to many reasons including the high cost of technology and lack of good facilities.
Transgenic animals: Transgenic animals are obtained when their hereditary DNA sequences are altered using different methods including addition of foreign DNA material or excision of undesired segment through recombinant DNA technologies. Furthermore, the technologies have enabled improvement of economically important traits such as resistance or tolerance to diseases and heat stresses. The major goals have been to obtain more productive animal.40,41 Unfortunately, this is also not achieved in Tanzania and other developing countries because of the significant costs of the technologies and lack of expertise. In addition there have concerns regarding acceptance of transgenic animals in many countries and it is likely that there might similar concerns if this technology is fully introduced in Tanzania.
Genetic markers and marker-assisted selection: Genetic markers enable the identification of DNA segments or genes which may influence expression of traits of interest by sequencing and screening genomes and carrying out association studies.42 Availability of large numbers of such markers has enhanced the likelihood of detection of major genes influencing quantitative traits. Marker assisted selection (MAS) is the process of selecting for a particular trait using genetic markers and this technique can accelerate the rate of genetic progress by increasing accuracy of selection thereby reducing generation intervals.43 Notably, the MAS is very useful for traits with low heritability and when the marker explains a larger proportion of genetic variance than does the economic trait.44 Positive examples of MAS include elimination of factors predisposing sheep to Listeriosis or Salmonellosis45 and identifying a gene for resistance to Boophilus microplus (cattle tick) in crossbreds of Hereford x Shorthorn cattle.4
Characterization of animal genetic resources: The importance of indigenous livestock breeds lies in their adaptation to local environments including lack of quality feeds, high temperatures and diseases. However, in developing countries including Tanzania some of livestock breeds are in danger of loss or being extinct due to indiscriminate crossbreeding with other breeds and lack of controlled mating. Characterization enables determination of genetic characteristics through evaluations of variations, relationships and unique attributes of indigenous livestock breeds or populations. In Tanzania population classification has been largely based on subjective data based on phenotypic characteristics and external morphological features (body size, coat colour, horn size and orientation) and information obtained from local communities (adaptation to different ecological and feed conditions) which may be not adequate and sometimes are misleading.6 Polymorphism in gene products such as enzymes, blood group systems and leukocyte antigens have traditionally been used for measuring genetic distance among livestock species
In the present time, these are being replaced by more powerful technologies such as the use of mitochondria DNA47,48 and molecular markers such as microsatellites and SNPs among others.49,50 In Tanzania a wide gene pool (a broad diversity of livestock breeds, strains or ecotypes) has not been fully exploited due to inadequate knowledge on their genetic distinctiveness. At present there are potentials for carrying out these technologies in the country. Previous examples include characterization of TSZ using random amplified polymorphic DNA (RAPD) and microsatellite markers4, #ref285 as well as evaluation of genetic diversity in SEA goats using microsatellite DNA markers.52 Recently Msalya et al.5 reported the genetic structure in three TSZ strains and Tanzanian Boran and Friesian breeds using genome-wide SNPs.5 In the study, three indigenous strains of TSZ namely Sukuma, Tarime, and Maasai showed low genetic differentiation (FST) ranging from 0.011 to 0.013 among them (Table 1). Boran and Friesian breeds showed greater values of FST than the TSZ strains. Furthermore, admixtures were observed among the TSZ animals and can be treated as one major cluster.5 Although TSZ are treated as different strains at present,6,51 it is currently difficult to define and genetically differentiate one strain of TSZ from another. However, the TSZ were differentiated from Boran and Friesian breeds (Figure 1).
Detection of diseases in livestock: The invention of Polymerase Chain Reaction (PCR) has enhanced detection of diseases in animals using the sensitivity of DNA. Examples include detection of bovine leukosis virus,53 documentation of differences between peste des petite ruminants (PPR) and rinderpest viruses in ruminants54ss as well as detection of bacteria in animal products. In Tanzania, sequences linked to Brucella (B.) abortus were detected in DNA samples isolated from milk using PCR (Figure 2). In the study, the prevalence of 16S-23S rRNA gene of abortus was 30%.55 In the same study, it was shown that the pathogenic Escherichia (E.) coli O157:H7 tested negative by PCR in the analyzed samples. Elsewhere Msalya et al.56 showed that two alleles of the prion protein gene (PRNP) modulated the expre ssion of messenger RNA in the medulla oblongata in cattle using standard curve method of real-time PCR (Figure 3) a situation which is linked to the occurrence of BSE in cattle.56 It is therefore possible to predict different diseases using biotechnological techniques
Animal feeds and nutrition improvement opportunities: Although fibrous feeds such as crop residues have low digestibility, they are potentially available to offer good nutritive value in ruminants by balancing nutrients through growth of microflora in the rumen using biotechnological methods. Different options for improving rumen fermentation and enhancing the nutritive value and utilization of agro-industrial by-products and other forages are reported in literature.57 One example is to utilize the molecular biology of Lactobacillus bacteria and other species through construction of novel genes encoding highly active fibre-degrading enzymes. These genes are then inserted into bacteria to aid the functioning of the rumen.58 The cleavage of genes and transfer into rumen bacteria also play role in reduction of methane production capability, decrease proteolytic and deamination activities, and increase the ability for microbial production of specific amino acids.59 Other available options are increasing the rate of breakdown high lignin resources by cleavage of hard carbon-carbon bonds using lignase enzyme60 and modifying genomes of plants to solve the constraints of limiting amino acids.61 Elsewhere, genetic engineering of plants to produce desired traits and reduce levels of effects of anti-nutritional factors (ANFs) has been achieved combined with conventional plant breeding methods. Availability of analytical and plant genome laboratories at SUA and MARI offers the possibility of conducting these and other studies in Tanzania and it is an opportunity for improving the efficiency of ruminant digestion and utilization of a wider range of feeds than is currently possible.
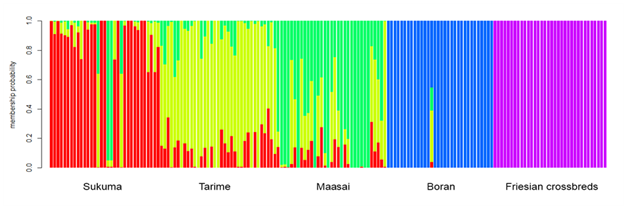
Figure 1 A structure map showing clusters among indigenous TSZ animals and previously imported breeds (Boran and Friesian) by membership probability at K=5 (Msalya et al., Unpublished).

Figure 2 Detection of 16S-23S rRNA sequences of B. Abortus in raw milk sampled from two regions of Tanzania.55
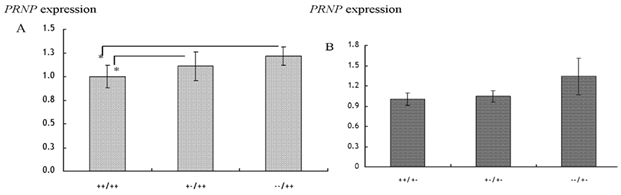
Figure 3 Expression of PRNP-mRNA (mean±SE) with respect to a 23-bp indel locus in A) Japanese black and B) Japanese brown cattle breeds.
*significance difference in the PRNP expression (p<0.05).56
Breed/Ecotype |
Tarime |
Maasai |
Boran |
Friesian |
Sukuma |
0.011 (±0.03) |
0.013 (±0.03) |
0.020 (±0.43) |
0.202 (±0.20) |
Tarime |
|
0.011 (±0.03) |
0.021 (±0.43) |
0.204 (±0.20) |
Maasai |
|
|
0.019 (±0.04) |
0.208(±0.20) |
Boran |
|
|
|
0.186 (±0.18) |
Table 1 Genetic variation (FST) among Tanzanian cattle (three strains of TSZ, Boran and Friesian)5
We have showed that there have been programmes to improve animal and animal feeds productivity in Tanzania for in the past years. Unfortunately, reports and success are difficult to find and show because most of these programmes were not sustainable and did not continue beyond programs’ or projects’ periods. Probably lack of good records, unclear objectives and sustainability strategies were some of the reasons of the failures of the past programmes in the livestock sector. This situation calls for researchers and implementers of existing and future programmes to come with strategies which will sustainably take the livestock an extra mile. The ongoing research and development programmes include the East Africa Dairy Development Project (EADD) aimed increasing ownership of improved breeds and increasing milk production among other objectives, the African Chicken Genetic Gains (ACGG) which tests the performance of five breeds/strains of chickens, and the Public-Private Partnership for Artificial Insemination Delivery (PAID) program which intends to establish more sustainable, efficient and effective private sector and government-led channels for delivering AI services and improve dairy cattle productivity. These programmes may improve livestock productivity if sustainability strategies will be in place. We have showed that different biotechnological techniques are now applicable and therefore existing and future projects can embrace the technologies for more significant results.
We declare that no conflict of interest exists for this article.

©2017 Msalya, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.