Journal of
eISSN: 2377-4312


Research Article Volume 8 Issue 5
1Bioenergetics and Environmental Sciences Division, ICAR-National Institute of Animal Nutrition and Physiology, India
2PhD Scholar, Department of Biochemistry, Jain University, India
3Knowledge management and Biostatistics Section, ICAR-National Institute of Animal Nutrition and Physiology, India;
4Animal Nutrition Division, ICAR-National Institute of Animal Nutrition and Physiology, India
Correspondence: Manpal Sridhar, Bioenergetics and Environmental Sciences Division, ICAR-National Institute of Animal Nutrition and Physiology, Adugodi, Bengaluru 560 030, India
Received: December 08, 2019 | Published: December 16, 2019
Citation: Rao RG, Ravichandran A, Kandalam G, et al. Enhanced production of manganese peroxidase from Clitopilus scyphoides employing statistical optimization for application in improving crop residue digestibility by ruminants. J Dairy Vet Anim Res. 2019;8(5):190-203 DOI: 10.15406/jdvar.2019.08.00267
Media components for the production of lignolytic enzyme MnP in the submerged fermentation from Clitopilus scyphoides were screened by PB design of experiments using main effects of different components and their percent contribution towards the MnP production. Total 31 components were considered in this study at temperature 30°C and incubation period of 5 days, in which Galactose, Malt extract, KH2PO4, NH4H2PO4, CaCO3, Tween80, Tartaric acid and sodium acetate were found to be significant components for MnP production. Further, response surface methodology was employed to optimize the media containing screened components along with temperature, pH and incubation days. The optimized medium contained 1.5% Galactose, 0.06%Malt extract, 0.02%KH2PO4, 0.15%NH4H2PO4, 0.003%CaCO3, 0.8%Tween80,0.005% MnSO4,0.88%Tartaric acid and 3% sodium acetate at temperature 31°C and pH 5.2 incubated for 5 days increased MnP productivity by 43 folds (43.2±1.29 U mL-1)when compared to un-optimized media (1.1±0.087 U mL-1). MnP produced was tested for lignolytic effects on different crop residues. Digestibility studies of different crop residues treated with crude MnP rich extracts indicated that a significant decrease in the percentage value of lignin and increase in in-vitro digestibility matter (at confidence interval 95%) when compared with untreated straws.
Keywords: Manganese peroxidase, Clitopilus scyphoides, crop residues, WRF, media optimization, RSM, PB design
Crop residues as straws symbolize as important renewable feed resource on earth providing energy for animal production in developingcountries like India, where these feedstuffs make up the main dietary component for animals.1 Crop residues are the fibrous by-products which result from the cultivation of cereals, pulses, oil plants, roots and tubers. These residues provide fodder at low cost since they are by-products of existing crop production activities. They are important adjuncts to natural pastures and planted forages and are often used to fill feed gaps during periods of acute shortage of other feed resources.2
When consideringforage digestion in rumen, higher lignin proportions are of concern due to reduced availability of crop cell wallpolysaccharides brought about by the presence of recalcitrant lignin. Biological treatment of agricultural residues is an emerging method for improvement of their digestibility which has fascinated many workers to work on the problem. As a result, utilization of white rot fungi (WRF) belonging to basidiomycetes have been extensively studied which selectively and efficaciously degrade lignin.3 This selective lignin degrading ability of white rot fungi is due to the presence of extracellular lignin modifying enzymes (LMEs) which consist mainly Laccase, Manganese peroxidase (MnP), Lignin peroxidase (LiP), and Versatile peroxidase (VP)along withoxidases which generate extracellular H2O2.4–6 Number of studies suggests that the lignolytic enzyme MnP produced from WRF is a more predominating enzyme causing and greatly contributing to degradation of lignin.7
Most researchers have worked on delignification of different crop residues by white rot fungi as well as for improving the protein status of crop residues by employing solid state fermentation.8–12 Though the solid-state fermentation on crop residue yielded positive result of delignification, severe loss of dry matter was observed and reported by many researchers.7,13 Treating of exogenous fibrolytic LMEs on crop residues to attack on lignin instead of direct fungal colonization on straws is advantageous for overcoming the problem of dry matter reduction.7,14
MnP is commonly produced by most of WRF in nature. But, limiting amounts of lignocellulolytic enzymes produced by WRF impede their commercial use. Many studies suggest that MnP production can be improved by screening and genetic manipulation of microorganisms, optimization of culture conditions and nutrients, development of new fermentative processes. The use of immobilization technique with media component optimization will help to overcome the limiting amount of enzyme secreted by the native fungus and enhance production.15 Medium composition and operation systems greatly affect the production of MnP. Carbon and Nitrogen sources are major chemical factors which play an important role in MnP production as in the fermentation of any other secondary metabolites by filamentous fungi.16 With medium optimization, researchers have reported MnP production as high as 70.20 UmL-1 in 8 days.17 Major media components like glucose, galactose, sucrose, lactose, glycerol, mannitol, maltose, Ammonium dihydrogen orthophosphate (NH4H2PO4), Ammonium sulfate, ammonium chloride, urea, malt extract, yeast extract, peptone, vitamins, amino acids ammonium tartrate, MnSO4, Tween 80,K2HPO4, KH2PO4, CaCl2, CaCO3 FeCl3, MgSO4 are used most often for the production of MnP.17–28
With an enormous biotechnological potential and wide variety of lignin degradation applications of MnP, this study was designed to a screen the significant factors form the pool of components to optimize the production of MnP by wildWRF isolate with the help of different statistical designs. Also, in this study, lignin degradation by the produced MnP enzyme in eight different crop residues commonly used for feeding ruminants was deliberated upon with the corresponding changes in the In-vitro dry matter digestibility.
Culture media, media components and analytical chemicals were purchased from Hi-Media, Mumbai, India. Chemicals used in the study were of analytical grade unless otherwise stated.Reactive Black 5 (RB5) and 2, 2’-azino-bis (3-ethyl benzothiazoline-6-sulfonic acid) (ABTS) used for assay were procured from Sigma Aldrich (USA). Polyurethane Foam (PUF) was procured as sheets from the local market and cut into 1cm3pieces.
Isolation, identification and phylogenetic analysis
Fruiting body of a mushroom on decaying wood from forest of Agumbe(13.5187 degrees N, 75.0905 degrees E) wascollected in a clean polythene self-sealing bag and labeled as AGUM004. Pure culture of collected sample was obtained by tissue culture technique.29–30 Then the pure culture was maintained on PDA slants and stored at 4°C for further use.
The collected culture was identified using ITS primer amplification of DNA and sequence blasting.31 Culture name was assigned based on more than 99% sequence similarity and the sequence was deposited at NCBI. In order to study the morphological characters of AGUM004, culture stained with lactophenol blue dyewasexamined under microscope. The morphology of the fungus and details of hyphae structure and spores were observed at 60x magnification. Phylogenetic study was obtained from MTCC, Chandigarh, India. The evolutionary history was inferred using the Neighbor-Joining method.32 The optimal tree with the sum of branch length = 0.28250559 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.33 The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood methodand are in the units of the number of base substitutions per site.34 The analysis involved 10 nucleotide sequences. All positions containing gaps and missing data were eliminated. There was a total of 549 positions in the final dataset. Evolutionary analyses were conducted in MEGA6.35
Screening for lignin modifying enzymes (LME)
Initial screening for lignolytic enzymes production was carried out on 0.02% Guaiacol supplemented LME basal agar medium which contains 0.1% KH2PO4, 0.05% C4H12N2O6, 0.05% MgSO4.7H2O, and 0.001% of CaCl2 and 0.01% Yeast extract, 0.0001% of CuSO4.5H2O, and Fe2(SO4)3,0.5% of Tween 80 and 0.002% MnSO4.H2O with 2% of Agar. Plates were inoculated with culture by means of standard agar plug technique in triplicates.29 The inoculated plates were incubated for 10 days and were checked for oxidation of guaiacol. Production of lignolytic enzymes were confirmed by the formation of halo reddish brown ring due to the oxidation of guaiacol. The culture was then screened for Manganese peroxidase production along with other lignolytic enzymes using LME basal liquid medium without agar. 50ml of LME basal medium in 250ml shaken flasks was inoculated with two40mm culture plugs from well grown culture on PDA plate for incubated 7 days at 30°C. Supernatant from the culture broth was used to check the production of MnP and other lignolytic enzymes such as Laccase, Lignin Peroxidase and versatile peroxidase.
LME quantification
Culture filtrate was used as the enzyme source to determine the ligninolytic enzymes activity. A sample of500 µL was taken to check different enzyme production. Laccase production was assessed by a measurement of the enzymatic oxidation of ABTS at 420 nm.36 The reaction mixture contained 0.8mM ABTS, 0.4 M sodium acetate buffer (pH5.2), and 500 µL of 0.5 mg/mL Catalase. The MnP activity was assayed by the oxidation of 4mM MnSO4 in 50mM sodium malonate buffer (pH 4.5), in the presence of 0.4mM H2O2. Oxidation of MnSO4 was measured by increase in OD at 270 nm.37 Lignin peroxidase was determined by Azure B assay.38 Reaction mixture for assay contained 0.04mM Azure B, 100mM sodium tartrate (pH 4.5), and 0.4mM H2O2. Oxidation of Azure B was determined by decrease in OD at 651nm. Production of versatile peroxidase was assessed by RB5 assay.5 Oxidation of RB5 was determined in 100mM sodium tartrate buffer at pH 3 with 10 µM of RB5. The reaction was initiated by addition of 0.1mM H2O2 and assessed through decrease in absorbance at 598 nm.
Screening of nutrient components for enhanced MnP production using Placket–Burman design
For inoculum, the fungal culture grown in growth medium, basically a LME basal medium without MnSO4 and Tween 80 were considered. The fungal cells grown in pellets were homogenized prior to inoculation and added to the production medium. Total 31 different Components were selected to check the main effect on production of MnP inoculated with 2% homogenized inoculum in 250ml shake flaks (duplicates) with 50ml media incubated for 5 days at 28°C and 120 rpm. Response was measured in terms of MnP activity (Uml-1) which was estimated by sodium malonate assay. The factors considered for the study were Linoleic acid, Vitamin solution, Suberic acid, MnSO4, lactic acid, Galactose, Glucose, Glycerol, Lactose, Maltose, Sucrose, Yeast Extract, Malt extract, Peptone, Urea, (NH4)2SO4, NH4H2PO4, K2HPO4, KH2PO4, MgSO4, CaCl2, FeCl3,CaCO3,CuSO4, Tween 80, Sodium Malonate, Tartaric acid, Succinic acid, Sodium acetate, Citric acid and Amino acid mixture (Table 1). A two-level factorial PB design used to evaluate the main effect was based on the first order model which can be explained by following equation (eq. 1)
(1)
|
S. No |
Factors in code |
Factors |
Units |
Minimum |
Maximum |
|
1 |
A |
linolaic acid |
g/L |
0 |
1 |
|
2 |
B |
Vitamin Solution |
% |
0 |
1 |
|
3 |
C |
suberic acid |
g/L |
0 |
1 |
|
4 |
D |
MnSO4 |
g/L |
0.01 |
0.03 |
|
5 |
E |
lactic acid |
g/L |
0 |
1 |
|
6 |
F |
Galactose |
g/L |
0.6 |
6 |
|
7 |
G |
glucose |
g/L |
0.6 |
6 |
|
8 |
H |
Glycerol |
g/L |
0.6 |
6 |
|
9 |
I |
Lactose |
g/L |
0.6 |
6 |
|
10 |
J |
Maltose |
g/L |
0.6 |
6 |
|
11 |
K |
Sucrose |
g/L |
0.6 |
6 |
|
12 |
L |
Yeast Extract |
g/L |
0.02 |
0.2 |
|
13 |
M |
malt extract |
g/L |
0.02 |
0.2 |
|
14 |
N |
Peptone |
g/L |
0.02 |
0.2 |
|
15 |
O |
NH4H2PO4 |
g/L |
0.1 |
0.8 |
|
16 |
P |
Ammonium sulfate |
g/L |
0.1 |
0.8 |
|
17 |
Q |
Urea |
g/L |
0.1 |
0.8 |
|
18 |
R |
K2HPO4 |
g/L |
0.1 |
0.6 |
|
19 |
S |
KH2PO4 |
g/L |
0.1 |
0.4 |
|
20 |
T |
MgSO4 |
g/L |
0.1 |
0.5 |
|
21 |
U |
CaCl2 |
g/L |
0 |
0.01 |
|
22 |
V |
Fecl3 |
g/L |
0 |
0.06 |
|
23 |
W |
CaCO3 |
g/L |
0 |
0.025 |
|
24 |
X |
CuSO4 |
g/L |
0.01 |
0.1 |
|
25 |
Y |
Tween 80 |
g/L |
0.25 |
1 |
|
26 |
Z |
Sodium Malonate |
g/L |
0 |
8 |
|
27 |
A’ |
Tartaric acid |
g/L |
0 |
8 |
|
28 |
B’ |
Succinic acid |
g/L |
0 |
1 |
|
29 |
C’ |
sodium acetate |
g/L |
0 |
1 |
|
30 |
D’ |
citric acid |
g/L |
0 |
1 |
|
31 |
E’ |
†amino acid mixture |
% |
0 |
0.1 |
Table 1 Factors and the range considered for PB experiment
Where Y is the response, β0 is the model intercept and βi is the variable estimate and X is the independent factors.39 The factors with positive effects with higher percent of contribution towards the response at 99% CI are considered most significant factors for MnP production. 31 factors were screened in 32 combinations each carried out in duplicate (Table 1).
† stock of amino acid mixture was prepared by dissolving 10mg of each glycine , cystine, aspartic acid, asparagine , arginine and histidine in 5oml of water.
Response surface methodology (RSM) and validation of statistical model
On the basis of PB experiment, factors screened as well as physical parameters like temperature, pH, PUF cubes for immobilizing the fungal culture and inoculum size (%) were selected to enhance the production of MnP. Galactose, Malt extract, KH2PO4, NH4H2PO4, CaCO3, Tween 80, MnSO4, Tartaric acid, sodium acetate, temperature, pH inoculum size, incubation days and PUF cubes were chosen as independent variables to optimize the production of MnP which is the dependent response variable at fixed orbital shaking of 80 rpm. The fungal cells grown in pellets in growth medium were homogenized prior to inoculation and added to the production medium as per the requirement. Independent variables with experimental range of an experimental design are shown in Table 2.
|
Factor |
Name |
Units |
Minimum |
Maximum |
|
A |
Incubation time |
Days |
3 |
7 |
|
B |
Temperature |
oC |
25 |
40 |
|
C |
pH |
3 |
7 |
|
|
D |
Inoculum Size |
% |
2.5 |
7 |
|
E |
PUF |
% |
2 |
6 |
|
F |
Galactose |
% |
0.5 |
1.5 |
|
G |
Malt extract |
% |
0.02 |
0.08 |
|
H |
NH4H2PO4 |
% |
0.04 |
0.15 |
|
J |
CaCO3 |
% |
0.001 |
0.005 |
|
K |
K2HPO4 |
% |
0.02 |
0.08 |
|
L |
Tween 80 |
% |
0.02 |
0.2 |
|
M |
Tartaric Acid |
% |
0.1 |
1 |
|
N |
sodium acetate |
% |
0.03 |
0.3 |
|
O |
MnSO4 |
g/L |
0.02 |
0.08 |
Table 2 Experimental range for individual factors used in Response surface methodology
Quadratic model in Central Composite Design (CCD) under Response Surface Methodology (RSM) was used to statistically optimize the adsorption capacity. The study design of 140 experiments along with experimental and predicted values is in agreement (Table 3). The model can be explained by following general equation
(1)
Where R represents the dependent response variable, β0 is regression coefficient; βi is the linear effect, βii is the squared effect and βij is the interaction effect of independent variable x. Design expert version 7.0 (stat ease, Inc., Minneapolis, USA) statistical software was used for RSM study and graphical representation (3D and contour plots) for the effect of independent variables on the response. The experiment was carried out in triplicates and values stated are mean values of obtained response (Table 3).
Validation and scale up of statistically optimized condition
MnP was biosynthesized by C. scyphoides using statistically optimized medium and conditions continuously for 12 cycles with repeated media replenishment for every 5 days in a 5L conical flask with 3L where culture was immobilized to the PUF initially added in the growth medium itself. MnP activity from all the harvests was calculated and the average activity from 12 cycles was compared with statistically projected value.
Study of In vitro digestibility of different crop residue using MnP
Crude enzyme produced from optimized media was then used to check the lignin degradation ability of MnP enzyme by proximate analysis40 and in-vitro digestibility analysis.41 Eight different straws were taken viz., Finger millet (Eleusinecoracana), Kodo millet(Paspalumscrobiculatum), Little millet (Panicumsumatrense), Proso millet (Panicummiliaceum), Brown top millet (Urochloa ramose), Barnyard millet (Echinochloaesculenta), Foxtail millet (setariaitalica) and Paddy (Oryza sativa). To evaluate the lignin degradation by enzymes, straws were powdered to approximately 1 cm and treated with enzymes in 1:2.5 (enzyme: straw) ratios and incubated for 24h at room temperature. The straws were dried at 60±5 ºC to stop the enzyme reaction. The treated straws were analyzed for DM (Dry Matter), ADF (Acid Detergent Fiber), NDF (Neutral detergent fiber), ADL (Acid Detergent Lignin), Protein and IVDMD (In-vitro dry matter digestibility) and compared with untreated strawsascontrol.42
Isolation, identification and Phylogenetic analysis
Isolated pure culture AGUM004 was identified as Clitopilus scyphoides of agaricales family. The sequence of 736 bp containing 18.5s rDNA, ITS1 and ITS2 region was submitted to GenBank with accession MH172163. Morphological details reveal that C. scyphoides had septate hyphae and sporulation was not evident (Figure 1). Cadogram (Figure 2) obtained from the phylogenetic analysis revealed that AGUM004 closely associated with genus Clitopilus (Figure 1&2).
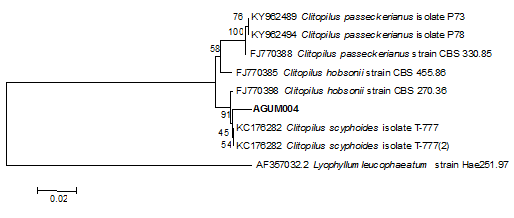
Figure 2 Cladogram of Clitopilus scyphoides revealing the phylogenetic relationship with other basidiomycetes.
Screening of MnP and other lignolytic enzymes by C. scyphoides
Initial screening of AGUM004on Guaiacol supplemented LME agar plates confirmed the secretion of lignolytic enzymes by the culture. Figure 3 clearly shows the oxidation of guaiacol by forming halo brown ring around the culture plugs (Figure 3).
Further investigation on lignolytic enzymes production by Clitopilus scyphoides revealed that, the WRF was a hyper MnP producer with 1110±100 U/L of MnP on seventh day of incubation. It wasalsorevealed that the culture produced 1300± 80U/Lof Laccase along with MnP. However,presence of LiP as well as VP was not detected in the culture filtrate.
Screening of important nutrient components for enhanced MnP production using Placket–Barman design
Since the culture was hyper MnP producer, PB was designed to screen the media component for enhanced production of MnP. All the 31 components considered for the production of MnP. The model is significant with F-value of 1398.49. R-square value of 99.9% the model is with 5.4% CV. The predicted R-squared value of 98.7% is in reasonable agreement with the adjusted R-Squared value of 99.8%.Table 4 shows the final screened factors which elucidated a positive effect on the production of MnP by the selected cultures. Total9 components were selected for C. scyphoides and for the optimization of MnP production based on their percent contribution, positive effect and p-value (≤ 0.01). Other factors which had negative effect, comparatively less percent of contribution and statistically not significant (P-value > 0.01) towards MnP production were rejected for the optimization study (Table 4).
|
Factors |
Coefficient |
% contribution |
F-value |
P-value |
|
MnSO4 |
2.31 |
2.63 |
28.80 |
0.003** |
|
Galactose |
15.18 |
3.97 |
43.35 |
0.0012** |
|
Malt extract |
15.60 |
4.19 |
45.76 |
0.0011** |
|
NH4H2PO4 |
28.17 |
13.66 |
149.32 |
0.001** |
|
K2HPO4 |
12.84 |
2.84 |
31.00 |
0.0026** |
|
CaCO3 |
19.12 |
6.29 |
68.80 |
0.0004** |
|
Tween 80 |
4.54 |
0.35 |
3.88 |
0.106 |
|
Tartaric acid |
11.72 |
2.36 |
25.83 |
0.0038** |
|
Sodium acetate |
19.64 |
6.64 |
72.60 |
0.0004** |
Table 4 Final screened factors from PB experiment for the optimization of MnP production
Response surface methodology (RSM) and validationof statistical model
Screened factors along with incubation time, inoculum size, pH and temperature were considered for response surface methodology to obtain optimized parameters for enhanced MnP production.Experiments were carried out with different combinations of 14 independent variables to study the individual as well as combined effects. Analysis of variance (Table 5) obtained from the quadratic regression analysis clearly shows the significance of individual and combined effect of these factors.
+ Suggestive significance (p value: 0.05<p<0.10) * moderately significant (p value: 0.01<p£ 0.05) ** strongly significant (p value: p£0.01)
Significance of factors were considered at confidence interval of 95% with p-value <0.05%. In this study A, B, C, D, F, G, N, O, AC, AK, BC, BE, BJ, BM, BN, CD, CM, DK, DL, DN, DO, EF, EM, EN, FO, GJ, GL, HM, A2, B2, C2, E2, F2, H2, and K2 are significant model terms and rest of the variables is insignificant. The RSM model is highly significant with model F-Value of 20.28. Insignificant lack of fit, high R2 value of 99.2%, adjusted R2 value of 94.3% and coefficient of variance (CV) of 17.7% assure that model can be used to navigate the design space. The regression equation obtained from the study is shown below equation (Eq-2)
MnP Activity UmL-1= -1793.7+99.7*A+76.3*B +221.1*C – 30.9*D–90.4*E+302.2*F-1393.6*G-184.12*H+1767.78* J-539.69*K-45.81*L-11.76* M-58.5 *N+937.97*O +0.0304 *AB+0.376* AC - 0.115 * AD + 0.102* AE + 0.059* AF +13.04*AG + 3.95*AH+121.36* AJ+14.86* AK-2.06 *AL - 0.400*AM + 2.40*AN+13.29*AO-0.13*BC + 0.004*BD + 0.060*BE + 0.066*BF - 3.86*BG + 1.80*BH - 62.34*BJ + 3.56*BK + 1.05*BL -0.30*BM + 1.84*BN - 2.56*BO -0.30*CD - 0.010*CE+ 0.77* CF+14.00*CG-2.75*CH + 92.25*CJ + 2.06*CK + 0.41*CL + 1.73*CM - 3.07*CN - 7.59*CO-0.025*DE +0.70 *DF +8.51*DG + 4.92* DH -119.28 *DJ - 23.48 *DK-5.92 *DL+0.24 *DM +3.94*DN-18.96 *DO-0.94*EF-2.58*EG-4.82*EH -35.80*EJ +15.02*EK+2.75 *EL *+1.48EM *-5.53EN +9.28*EO + 30.96*FG-14.05*FH-545.46 *FJ -50.98*FK -6.92*FL -1.95*FM+12.38 *FN-85.16 *FO - 139.37*GH +17089.66*GJ+955.74 *GK+564.76 *GL+45.66 *GM+0.71 *GN-193.4 *GO+1008.72*HJ + 71.12 *HK-55.02 *HL+82.58 *HM +56.66*HN-209.62 *HO + 10415.8*JK + 5028.32*JL + 334.44*JM - 435.27*JN -9460.82*JO + 219.97 * KL+2.79 *KM -226.19*KN - 104.63*KO +7.28*LM + 34.40*LN +101.11*LO+2.58 *MN-32.32 *MO - 41.14*NO - 1.28* A2 - 0.12 * B2 -1.90*C2 -0.02*D2 0.84*E2 -9.24*F2 -360.06*G2 +550.68*H2 -202801*J2 + 3500.44*K2 -83.18*L2 0.19*M2 +27.43*N2 -5815.93*O2 (2)
The optimal values of the variables determined by maximization of the second-order polynomial equation with interaction terms obtained by multiple regression analysis based on CCD. Maximum adsorption obtained by the statistical optimization experiment was 55.5Uml-1 with optimized conditions established as a pH of 5.2, Galactose of 1.5%, NH2H4PO4 0.15%, 0.06% malt extract, 0.02% K2HPO4, 0.003% CaCO3, 0.08% of tween 80,0.88% tartaric acid 0.3% Sodium acetate and 0.005% of MnSO4for an incubation time of5 days with constant orbital shaking of 80 rpm at temperature of 31oC for 6.8% of inoculum and 2% PUF to immobilize the cells. For the production of MnP, incubationtime, pH, temperature, concentration of MnSO4 and Tween 80 have a partial positive effect on production of MnP. These factors show positive effect on MnP up to certain level of increase and beyond which show negative effect. But galactose, malt extract, NH4H2PO4, tartaric acid, and sodium acetate have shown positive effects on MnP Production. Increase in theses factor increases the activity of MnP directly. PUF and CaCO3 however have not shown direct effect on the production of MnP. The effect of few parameters in a pair on the MnP activity is graphically extrapolated in the form of 3D surface and contourplots (Figure 5–12). Graphs for MnP activity plotted with Incubation time VspH (Figure 5) clearly indicate that as number of days increased from zero to 7, MnP activity increases till 6th day and beyond that it starts decreasing and increase in pH from zero to 6 has positive effect whereas increase of pH above 6 is deteriorating for MnP. Even temperature also had exhibited similar effect on MnP production (Figure 6) showing thetemperature around 30oC as ideal for MnP production. Though malt extract has a positive effect on MnP production, the graph plotted for malt extract Vs incubation time (Figure 7) indicated that longer period of incubation has a negative effect on MnP even when the concentration of malt extract is maximum. Graph plotted for galactose Vs pH (Figure 8) depicted that increase in Galactose concentration has a positive effect but increase in pH beyond 6 has a negative effect for the production of MnP even when Galactose concentration is increased to maximum. Though MnSO4 is essential for the production of MnP, increase in the concentration beyond 0.05gL-1 shown no significant effect. Malt extract and Sodium acetate both have shown positive effect on the MnP production (Figure 9). Figure 10 in the graph plotted for Galactose Vs PUF shows that Galactose has a positive effect on MnP production whereas increase in PUF concentration had not contributed for the increased MnP biosynthesis. Graph plotted for MnSO4Vs inoculum size (Figure 11) depicted the positive effect of increased % of inoculum volume and the concentration of MnSO4. It is evident that even with increased inoculum size to maximum has no positive effect with the concentration of MnSO4 beyond 0.05gL-1. Figure 12 shows NH4H2PO4 concentration has positive effect on MnP Production. Even with lower concentration of Tween 80, by increasing concentration of NH4H2PO4, increased the activity of MnP. concentration of Tween 80 from 0.04% to 0.08% has proven to be positive and beyond 0.08% has no significant effect on MnP production Figure 4–12.

Figure 6 Contour and 3D response surface graph plotted for MnP activity with Incubation time VsMalt extract.
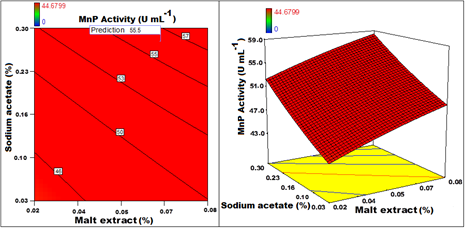
Figure 8 Contour and 3D response surface graph plotted for MnP activity with Malt extract Vs Sodium acetate.

Figure 10 Contour and 3D response surface graph plotted for MnP activity with Inoculum sizeVs MnSO4.
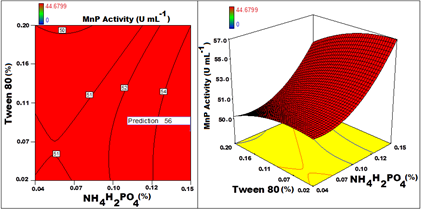
Figure 11 Contour and 3D response surface graph plotted for MnP activity with NH4H2PO4Vs Tween 80. The optimized predicted valueobtained from RSM for enhancing MnP production wasscaled up to 3L from 50mL. Process was validation with immobilized cells in the optimized medium (Figure 12).

Figure 12 Flask showing immobilized cells in optimized medium for the production of MnP from C.scyphoides.
Figure 14 shows the plot for activity of MnP from immobilized cells in submerged fermentation continuously for 12 cycles. The MnP activity obtained was 43.2±1.29 U mL-1 against 55.5 U mL-1 of MnP statistically predicted. Experimental yield of MnP was 77.7% when compared to predicted value. Production of laccase in this experiment was estimated to be 10.9± 1.6 U mL-1 (Table 6) (Figure 14).
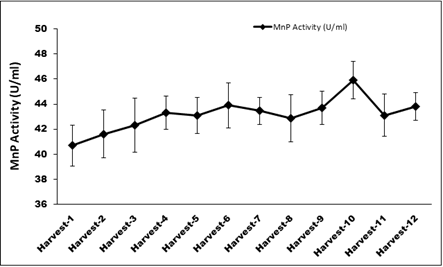
Figure 13 Continues MnP production through immobilized cells in optimized condition for 12 cycles.
+ Suggestive significance (p value 0.05<p<0.10)* moderately significant (p value 0.01<p£ 0.05) ** strongly significant (p value p£0.01).
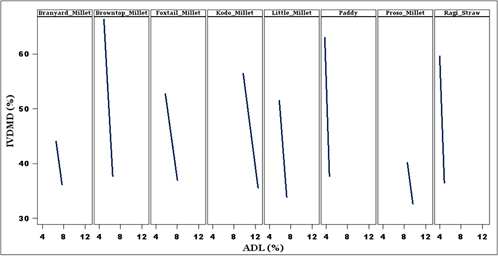
Figure 14 Regression Graph showing correlation between ADL (%) and IVDMD (%) for different straws treated.
Proximate analysis of all the straws treated with culture filtrate of C. scyphoides, revealed that there is a significant change in the percentage values of NDF, ADL, and ADF (at confidence interval 95%) when compared with control straws. All straws showed decrease in NDF, ADF and ADL. In-vitro digestibility of straws treated with MnP enzyme revealed that, in all the straws there is increase in % digestibility when compared with untreated control straws (Table 6) Finger millet and paddy straws were turned out be statistically promising results while straw and barnyard millet showed least response. Negative correlation was observed for correlation plots (Figure 15) plotted with ADL versus IVDMD with correlation coefficient of -0.83 for paddy, -0.92 little millet, -0.93 for foxtail millet, -0.81% barnyard millet, -0.97 for proso millet, -0.75 for brown top millet, -0.92 for finger millet and -0.81for kodo millet.
WRF are ubiquitous in nature and grow as saprophytes on dead and decaying trees usually in the forest ecosystem and are exceptional with the ability to selectively degrade lignin with the help of some extracellular enzymes like Laccase, MnP, LiP and VPalong with oxidases which generate extracellular H2O2. Basidiocarp of AGUM004 was collected from the forest where the substrate was represented by decaying wood which was the initial indication of presence of one or more LMEs. Further microscopic observation of spores, hyphae and septa was considered to determine it as basidiomycetes. As confirmatory test, molecular identification techniques involving DNA sequences which are more precise and efficient was considered to check whether collected wild isolate was member of basidiomycetes. Accordingly, more frequently employed method of using Internal Transcribed Sequence (ITS) region of ribosomal DNA through the use of ITS 1 and ITS 4 primers to identify unknown fungal isolate was adopted. The isolate was identified through comparison with BLAST of the known fungal sequences in a distinct study carried out in our laboratory. The phylogenetic analysis confirms the novel isolate belongs to Clitopilus genus which is a member of basidiomycetes.
Guaiacol is most widely used indicator compound to screen LME producing WRF when compared to other compounds like tannic acid, gallic acid, syringaldehyde, etc., which directly indicates the presence of LME.43 Though primary screening does not confirm the presence of any particular enzyme, it is easy and reliable to screen LME producing microorganisms.Naturally WRF produces more than one LME or array of LMEs to efficiently degrade lignin in the wood. Therefore, quantitative estimation of LME was carried out using LME basal medium. Quantitative analysis of culture filtrate by isolated culture revealed that AGUM004 is producer of Laccase as well as MnP. The quantitative analysis of the ligninolytic enzymes secreted confirms the existence of MnP.
MnP is an extracellular enzyme produced by WRF as a secondary metabolite and MnP is produced in very less amount in their native states. Moreover, production of LME always depends on growth condition of fungus as well as medium composition on which fungus is growing. Hence, we employed statistical methods to screen most significant components to increase the MnP production as well as to optimize a condition in which we can get maximum MnP yield through submerged fermentation technique. 31 factors selected for screening through placket barman design are considered as different carbon sources, nitrogen sources, inducers and cofactors based on previous studies on MnP production.22,18–20,24–28 The screened factors consisted of Galactose as sole carbohydrate source, malt extract and NH4H2PO4 as nitrogen source, Tween 80, MnSO4, sodium acetate and tartaric acid as inducers Screened factors along with factors responsible for optimum cultural growth like temperature, pH, inoculum percentage and PUF cubes to immobilize the fungal cells were also considered. Incubation temperature is very important factor for fungal growth as well as MnP production. Optimum temperature of 31 oC obtained is in agreement with other studies which have been reported that temperature in the range of 25 to 37 oC is optimal for the production of LME by various WRF.44–46 Initial pH of medium for the production of MnP is important as it is essential for the growth of microorganisms as well as metabolic action.47 Mostly reported range of pH for optimal growth of different WRF is between 4.5 to 6.5 and is similar to our finding for C. scyphoides.25,48 Studies have reported that LME synthesis depends on the carbon nitrogen ratio and nitrogen limiting medium favors the biosynthesis of LME.24,49,50 Our study revealed that for C. scyphoides, increase in nitrogen concentration has not shown any negative effect on the production of MnP. Production of secondary metabolites like enzymes depends on the time of incubation as it affects the growth and biosynthesis of metabolites.51,52 Optimum incubation time for C. scyphoides in our studies for maximum MnP production was found to be 5 days and MnP activity started decreasing after 6 days. Previous studies have also reported similar findings.17,46,53 Apart from physical factors and carbon, nitrogen sources, use of MnSO4, tween 80 and organic acids have proven inducer for MnP biosynthesis in different WR.18–20,28 Submerged production of MnP using statistically optimized conditions drastically increased the production of MnP in C. scyphoides. Production of Laccase did not drastically increase as MnP, in the media used. This can be explained by the requisite of copper ions supplementation in the media for the production of higher level of Laccase.54
77.7% yield of MnP by using statistical optimized condition indicates that the experimentally determined production values were in close agreement with the statistically predicted ones. The validation experiment thus confirms the reliability of the model. Digestibility studies of different crop residues treated with crude MnP rich extracts indicate that a significant decrease in the percentage values of NDF, ADL, and ADF and increase in IVDMD (at confidence interval 95%) when compared with untreated straws. MnP from C. scyphoides indeed helps in the degradation of recalcitrant lignin in turns increases the digestibility of otherwise low digestible crop residues by 15-20 units. Digestibility was found to be negatively correlated with lignin content which indicates the delignification of treated straws.
Wild isolate Clitopilus scyphoides was found to be Hyper MnP and laccase producer. PB design was employed to screen 31 factors selected based on previous studies on MnP production.Nine out of 31 factors (Galactose,malt extract, NH4H2PO4, K2HPO4, MnSO4, CaCO3, Sodium acetate, tartaric acid, tween 80) were screened through PB design. Nine factors along with physical parameters further considered for optimization of medium to maximize the MnP production using RSM. The statistically optimized condition was then scaled up and validated. MnP production was increased by 40 folds with optimized medium as compared to optimized medium. Thus, the optimized condition can be utilized for large scale production of MnP. Digestibility studies of different straws suggest that crude MnP from C.scyphoides indeed helps in the degradation of recalcitrant lignin in turns increases the digestibility of otherwise low digestible crop residues. Therefore, treatment of straws with MnP is recommended prior to feeding the ruminants.
The financial assistance from the Department of Biotechnology, Ministry of Science and Technology, Government of India (Grant No.BT/PR11205/AAQ/1/589/2014) to conduct the present research is gratefully acknowledged. The authors thank the Director of ICAR National Institute of Animal Nutrition and Physiology, Bangalore (India) for providing facilities to carry out the research work.
Author declares that there are no conflicts of interests.

©2019 Rao, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.