Journal of
eISSN: 2373-633X


Research Article Volume 8 Issue 2
1SASTRA University, India
2Purdue University, USA
Correspondence: Rahul Ravichandran, School of Chemical and Biotechnology, SASTRA University, Tamil Nadu, India
Received: April 28, 2017 | Published: June 12, 2017
Citation: Ravichandran R, Sundararajan R. In silico-based virtual drug screening and molecular docking analysis of phyto chemical derived compounds and FDA approved drugs against BRCA1 Receptor. J Cancer Prev Curr Res. 2017;8(2):216-220. DOI: 10.15406/jcpcr.2017.08.00268
Background: Molecular docking is used to identify natural photochemical and commercial drugs that are equivalent to treat BRCA1-based breast cancers. BRCA1 type breast cancers have high mortality and recurrence rate. BRCA1 gene which is also known as Breast Cancer Susceptibility gene 1, BRCA1 gene has high probability of mutation or duplication which further leads to breast cancer.
Materials and Methods: There are numerous photochemical and chemical drugs that are administered against breast cancer. These compounds are taken from the literature survey and they are meant to drug screening process against BRCA1 receptor (PDBID-4Y2G) using iGEMDOCKv2.1 and further the binding site of the Receptor is determined and the top 5 molecules that were attained from both phytochemicals and commercial drugs are proceeded to Active Site docking using Autodock vina software.
Results: Based on their Drug Screening scores and Binding affinity scores, one best phytochemical Epi Gallo Catechin Gallate-EGCG (Green tea extract), and one commercial drug, Doxorubicin Hydrochloride with best energies from docking are obtained.
Keywords:molecular docking, BRCA1, drug screening, EGCG, doxorubicin hydrochloride
Computer-Aided-Drug-Discovery approaches have been extensively used in drug investigation to increase the efficiency of the drug discovery and development pipeline, subject to the purpose and systems of interest 1. Docking is a software technique that fits a molecule into target binding sites. It is used for binding modes of protein with ligands and inhibitors 2. It is used to predict the structure of the intermolecular complex formed between two or more molecules. Thus, it is used for drug discovery and matching molecules, with overall minimum energy. In this research, the equivalent natural phytochemical and commercial drugs are identified using molecular docking for treating BRCA1 breast cancer.
The BRCA1gene is situated on the long arm of Chromosome 17 at 17q21 and BRCA1 also contains 24 coding exons range over 80 kb.3 BRCA1 was first mapped and was consequently cloned in the years 1990 and 1994.4,5 BRCA1 interacts with quite a lot of proteins that are involved in cell cycle progression, cellular pathways, ubiquitination, gene transcription regulation and DNA damage response.6 BRCA1 is a pleiotropic DNA Damage Response protein (DDR) that functions in both DNA activation and checkpoint repair.7 BRCA1 can bind to RAD51, p53, RAD50-MRE11-NBS1 (R-M-N) complex and leading to homologous recombination or Non-Homologous End-Joining (NHEJ) which is having excessive importance in DNA damage and its repair.8 Deficiency in BRCA1 as a result causes abnormalities in the centrosome duplication, the G2/M checkpoint, the spindle checkpoint and S-phase checkpoint.9 One of the most important features of BRCA1 associated breast cancer is that it comprises a complex unit of aneuploidy than tumours without Breast Cancer Susceptibility gene 1 mutations.10 HCC1937 cell line derived from homozygous BRCA1 deficient breast tumour shows a high number of chromosomal gains and losses, heterozygosity loss at numerous loci identified to be involved in the development of breast cancer.11
Deficient cells in BRCA1 revealed centrosome amplification, abnormalities in all most important cell cycle checkpoints and impaired DNA damage repair.1,12 Molecular docking has turned out to be an ever more important tool in the case of drug discovery. The main objective of molecular docking is to contribute a prediction of the ligand-receptor complex structure by means of computational approaches.13 Docking can be used to execute virtual screening on enormous libraries of compounds, rank the results based on the scores and put forward structural hypothetical theories on how the ligands inhibit the target receptor, which is vital in lead optimization.14 Factors affecting docking are: intramolecular (bond width, bond angle and dihedral angle) and intermolecular (electrostatic, dipolar, H-bonding, hydrophobicity and van der Waals) forces.
Primary sequence retrieval
The amino acid Fasta sequence of BRCA1 gene has been obtained from Uniprot database with ID (UniProtKB - P38398 (BRCA1_HUMAN)).15 This fasta sequence was used for further investigation.
Protein structure retrieval
The fasta sequence of BRCA1 was subjected to sequence similarity search using BLASTP16 and the one with 100% sequence identity structure was retrieved from Brookhaven RCSB PDB database with (PDBID-4Y2G).17 This PDB structure was used for further analysis.
Ligand archive research
Literature survey lead to the identification of 18 natural compounds that are belonging to Flavonoids, Isoflavones, Catechine, polyphenolic and terpenophenolic families18 and 26 Chemical drugs approved by the Food and Drug Administration (FDA) for Breast Cancer treatment.19 The 3D structures of these natural compounds and chemical drugs are downloaded from the PubChem database20 in SDF format and they are converted to the PDB format using Open Babel version 2.3.2 software.21
Drug screening using iGEMDOCK
All the 17 phytochemicals and the 26 chemical compounds are subjected to Drug Screening process against BRCA1 receptor (PDBID-4Y2G) using iGEMDOCKv2.122 a standalone virtual screening software The Drug Screening module is set with the Default parameters such as Population Size = 200, Generations = 70, Number of solutions =3. Out of three solutions the one with the least energy values are considered.
Active site prediction
Active Site region of the BRCA1 gene is predicted using CASTp webserver23 which gives the residues that are participating in the active site of the macromolecule including the active site’s area and volume. The active site residues are further mapped on to the structure to check exactly where the residues are present.
Molecular docking studies
Molecular Docking is a technique used to analyze the position and the inhibition interaction between the protein and the small molecule. Docking was performed using Auto Dock Vina24 which is standalone software. As a preprocessing step, The PDB format of Macromolecule and SDF format of small molecules are converted to Atudock’s PDBQT format. From the Auto Dock Vina the macromolecule was added with a Gasteiger Partial charges and further Hydrogens were added. The docking of small molecule to the macromolecule was focused on the specified binding site. The total number of rotatable bonds of the lignads is calculated. The Grid was defined to the Binding site of the protein structure with the configurations of x/y/z coordinates was set to size_x=62, size_y=84, size_z=60 centre of the grid box was set to center_x=-1.141, center_y=2.034, center_z=-24.991 in X,Y,Z dimensions, in which the grid was covered to the binding site of the macromolecule. The Binding energies between receptor and the ligands are attained in terms of Kcal/mol.
Protein-ligand interactions
The interactions like Hydrogen Bonding and other non-bonded terms between the top docked Phytochemicals, Commercial drug and the receptor are seen using Accelrys Discovery Studio Visualizer software 25. Figure 1 shows the methodology.
Protein structure visualization
The protein structure retrieved from RCSB PDB with (PDBID-4Y2G) was visualized using UCSF chimera which is molecular visualization software.26 The bound ligands are already removed and the structure without bound ligand has been taken for the in silico analysis. Figure 2 shows BRCA1 susceptibility protein structure visualisation using Chimera 1.10.2 software. BRCA1 structure has double chain such as chain-A represented in blue colour, chain-B represented in white with various atom representing various colours and its N-Terminal indicating starting point of the structure and C-Terminal indicating the end point of the structure.
Residues involved in active sites
The active site of the macromolecule BRCA1 has been determined using CASTp webserver. The determination of residues that are involved in the active sites are taken from Pocket number27 with an Area of 443Angstrom2, Volume of 701.4 Angstrom3 and the active site residues are shaded in Green colour as shown in Figure 3.
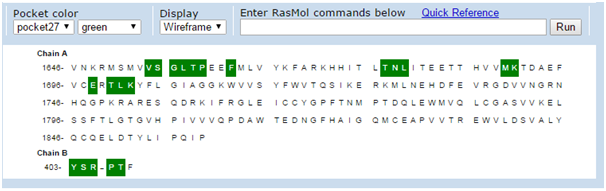
Figure 3 Shaded Regions representing active site residues using CASTp. Various residues from both the chains-A,B are involved. These residues are further mapped onto the structure to know where exactly the active site is present in the molecule.
Drug screeening using iGEMDOCKv2.1
As the phytochemicals and commercial drugs are subjected to drug screening, the top five small molecules of both Phytochemicals and commercial drugs were short-listed based upon their scores and they are subjected to further molecular docking studies. The features of the top five drug Screened compounds are summarized in Table 2. The virtual drug screening scores for these top five phytochemicals and commercial drugs were obtaned from iGEMDOCK. The highly negative energy score represents the stronger binding affinity between the receptor and the small molecule, whereas positive score represents to poor or no effective binding. Curcumin (turmeric extract), Lapatinib Ditosylate (commercial drug) attained the highest score.
Phytochemicals |
Pubchem ID |
Commercial Drugs |
Pubchem ID |
Apigenin |
Pubchem CID-5280443 |
5-Fluoro Uracil |
Pubchem CID-3385 |
Biochainin A |
Pubchem CID-5280373 |
Abitrexate |
Pubchem CID-126941 |
Carotenoids |
Pubchem CID-16061280 |
Abraxane |
Pubchem CID-36314 |
CBD |
Pubchem CID-26346 |
Anastrozole |
Pubchem CID-2187 |
Curcumin |
Pubchem CID-969516 |
Aredia |
Pubchem CID-73351 |
Cyanidin |
Pubchem CID- 68247 |
Aromasin |
Pubchem CID-60918 |
Daidzein |
Pubchem CID-5281708 |
Capecitabine |
Pubchem CID-60953 |
EGCG |
Pubchem CID- 65064 |
Clafen |
Pubchem CID-2907 |
Gabridin |
Pubchem CID-124052 |
Docetaxel |
Pubchem CID-148124 |
Genistein |
Pubchem CID-5281377 |
Doxorubicin Hydrochloride |
Pubchem CID-443939 |
Hesperitin |
Pubchem CID-72281 |
Ellence |
Pubchem CID-41867 |
Ipriflavone |
Pubchem CID-3747 |
Eribulin Mesylate |
Pubchem CID-17755248 |
Kaempferol |
Pubchem CID-5280863 |
Everolimus |
Pubchem CID-6442177 |
Lignans |
Pubchem CID-261166 |
Fareston |
Pubchem CID-3005573 |
Luteolin |
Pubchem CID-5280445 |
Faslodex |
Pubchem CID-104741 |
Naringenin |
Pubchem CID-439246 |
Femara |
Pubchem CID-3902 |
Quercetin |
Pubchem CID-5280343 |
Gemicitabine Hydrochloride |
Pubchem CID-60749 |
Resveratrol |
Pubchem CID-445154 |
Goseralin Acetate |
Pubchem CID-16052011 |
Ibrance |
Pubchem CID-5330286 |
||
Ixabepilone |
Pubchem CID-6445540 |
||
Lapatinib Ditosylate |
Pubchem CID-9941095 |
||
Megestrol Acetate |
Pubchem CID-11683 |
||
Nolvadex |
Pubchem CID-2733526 |
||
Thiotepa |
Pubchem CID-5453 |
||
Trastuzumab |
Pubchem CID-3001322 |
||
Velban |
Pubchem CID-46783244 |
Table 1 The 18 compounds of phyto chemicals and the 26 compounds from commercial drug list that were studied
Name of receptor |
Natural compound |
iGEMDOCK energy (phytochemicals) |
Commercial drug |
iGEMDOCK energy (commercial drugs) |
BRCA1 (4Y2G) |
Curcumin |
-101.5 |
Lapatinib Ditosylate |
-115.6 |
BRCA1 (4Y2G) |
EGCG |
-99.9 |
Doxorubicin Hydrochloride |
-113.1 |
BRCA1 (4Y2G) |
Apigenin |
-95 |
Ellence |
-109.2 |
BRCA1 (4Y2G) |
Quercetin |
-93.9 |
Abitrexate |
-105.5 |
BRCA1 (4Y2G) |
Kaempferol |
-93.6 |
Trastuzumab |
-105.1 |
Table 2 Summary of Top-5 Phytochemicals and Commercial Drugs Virtual Drug Screening Scores obtaned from iGEMDOCK
Molecular docking
Molecular Docking was done taking these top five small molecules from both phytohemicals and commercial drugs and further the molecules were docked to the active site of the protein. Various binding affinities are obtained for all the ligands in terms of (Kcal/mol). The various features and the binding affinities of the docked complexes of are summarized in Table 3, which shows the summary of Top five Phytochemicals and Commercial Drugs binding affinity scores obatined from AutoDockvina interms of kcal/mol. The highly negative energy score represents the stronger binding affinity between the receptor whereas EGCG (green tea extract) and Doxorubicin Hydrochloride attained the highest binding affinities from the top five screened compounds. Table 4 shows the chemical structure of the top 2 Phytochemicals and Commercial Drugs Binding Afiinity scores obatined from iGEMDOCK and AutoDockvina.
Name of the receptor |
natural compound |
Autodock energy Kcal/mol |
Commercial compound |
Autodock energy Kcal/mol |
BRCA1 (4Y2G) |
Curcumin |
-6.1 |
Lapatinib Ditosylate |
-7.5 |
BRCA1 (4Y2G) |
EGCG |
-7.9 |
Doxorubicin Hydrochloride |
-8.1 |
BRCA1 (4Y2G) |
Apigenin |
-7.1 |
Ellence |
-8 |
BRCA1 (4Y2G) |
Quercetin |
-6.6 |
Abitrexate |
-6.1 |
BRCA1 (4Y2G) |
Kaempferol |
-6.7 |
Trastuzumab |
-7.6 |
Table 3 Summary of Top-5 Phytochemicals and Commercial Drugs Binding Afiinity scores obatined from Auto Dockvina
BRCA1-EGCG complex
Figure 4 shows the docked complex of BRCA1 receptor. Dark blue represents chain-A and white represents chain-B whereas light blue represents EGCG compound in Sticks model highlighted in the box. This docked confirmation of EGCG with BRCA1 has obtained the highest binding affninty of -7.9 Kcal/mol using Autodock vina.
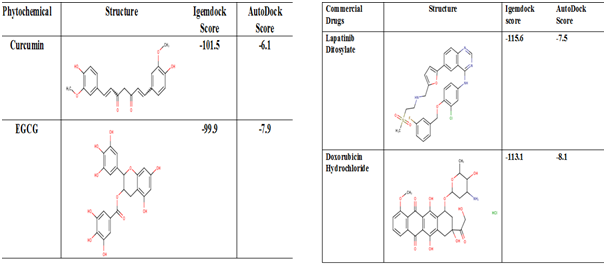
Figure 4 Summary of Top-2 Phytochemicals and Commercial Drugs Binding Afiinity scores obatined from iGEMDOCK and AutoDockvina.
Ligand plot for residual wise interaction
Figure 5 shows BRCA1 Protein and EGCG Receptor interaction plot viusalised using Discovery studio viusalizer. It represents various residues corresponding to different types of interactions of BRCA1 with EGCG. A conventional hydrogen bond is seen between Leucine:1657 (A-chain) of BRCA1 and the EGCG molecule and other non-bonded interacting residues can be seen.
4Y2G doxorubicin hydrochloride complex
Figure 6 shows the docked complex of BRCA1 receptor. Here, dark blue represents chain-A, white represents chain-B and light blue represents Doxorubicin Hydrochloride in Sticks model highlighted in the box. This docked confirmation of Doxorubicin Hydrochloride with BRCA1 has been obtained with the highest binding affinity of -8.1Kcal/mol using Autodock vina.
Ligand plot for residual wise interaction
Figure 7 shows BRCA1 Protein and Doxorubicin Hydrcholoride drug interaction plot visualised using Discovery studio visualizer. It represents various residues corresponding to different types of interactions of BRCA1 with Doxorubicin Hydrochloride. A conventional hydrogen bond is seen between Proline 407 (B-chain) of BRCA1 and the Doxorubicin Hydrochloride molecule and other non-bonded interacting residues can be seen.
It takes years and millions of dollars to discover a drug and bring to real world use. To reduce the cost and time, it is of practical interest to study the drug molecules and their characteristics using commercial/industrial software. Drugs interact with receptors in a highly specific and complimentary manner. Towards this, in this study, we compared the naturally occurring phytochemical compounds and commercial drugs which are similar in characteristics for BRCA1 type breast cancers. Breast cancer was considered as it is the most common type of cancer in women worldwide. Each year over 1 million new cases occur and over 500,000 die. For the patients on chemo therapy, side effects and drug cost add to the physical suffering due to the cancer. Hence, it is critical that we identify natural compounds which have least or no side effects and are also affordable.
The current proposed hypothesis deals with the Virtual Screening & molecular docking procedures for different small molecules against BRCA1 protein structure for checking its binding affinities and inhibitory activity of small molecules against the receptor. The screened compounds such as EGCG (Phytochemical) & Doxorubicin Hydrochloride (commercial drug) has shown the best binding affinity towards the BRCA1 receptor and EGCG’s Energy is -7.9kcal/mol whereas Doxorubicin Hydrochloride’s energy is -8.1kcal/mol as both the compounds have very slight difference in their binding affinity scores can be hypothesized that the Phytochemicals can also be considered as an inhibitor against Breast cancer susceptibility gene 1 for breast cancer treatment apart from these commercial drugs. Further experimental studies are needed to understand the mechanism of action of phytochemicals against Breast cancer.
None.
The authors declare that there are no conflicts of interest.
None.

©2017 Ravichandran, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.