Journal of
eISSN: 2469 - 2786


Research Article Volume 5 Issue 3
Department of Biotechnology, Cochin University of Science and Technology, India
Correspondence: Padma Nambisan, Department of Biotechnology, Cochin University of Science and Technology, Cochin- 682022, Kerala, India, Tel +919446535151
Received: July 20, 2017 | Published: August 31, 2017
Citation: Lakshmi KMS, Soumya PS, Shaji A, et al. Lenzites elegans KSG32: a novel white rot fungus for synthetic dye decolourization. J Bacteriol Mycol Open Access. 2017;5(3):311-317. DOI: 10.15406/jbmoa.2017.05.00138
Lenzites elegans, a ligninolytic enzyme producing white rot fungus with proficient synthetic dye decolourization ability was isolated from decaying wood. To the extent of our knowledge, this is the first report of laccase (362.7IU/mL) and lignin peroxidase (5.17IU/mL) production by L. elegans KSG32. The synthetic dye decolorizing ability of the fungus was checked using solid and liquid media. The organism showed greater ability for decolourization of naphthol green B (97%) and bromophenol blue (88%) in the submerged condition followed by partial decolourization for two azo dyes, namely, orange G (70%) and congo red (62%). This is an indicative of the potential application of L. elegans KSG32 in decolourization of colored industrial effluent. The FTIR analysis of the decolourized dye sample showed that major bonds in the dye were broken and oxidation process also increased. The analysis of biomass content showed no growth inhibition of L. elegans KSG32 in the presence of dyes, indicating the less toxicity of these dyes on fungal growth.
Keywords: ligninolytic fungi, laccase, lignin peroxidase, synthetic dyes, decolorization, FTIR
Synthetic dyes are complex aromatic compounds used in many industries like paper, printing, textile, food and cosmetics.1 The major synthetic dyes include azo, triphenylmethane, anthraquinone and metal dyes. They are reported to be carcinogenic and thus their safe release into the environment poses a threat to public health. About 10-15% of the substrate unbound dyes from the industries are discharged into water bodies without any effective treatment.2 The release of these effluents into water bodies affects all biological systems.3 The removal or degradation of these dyes can be done by both physical and chemical methods. But the production of secondary pollutants like sludge and toxic compounds discourages the use of such costly methods.4 These disadvantages can be overcome by adopting biological methods or green strategies. The most cost-effective and eco-friendly approach is the removal of these dyes by various microorganisms and associated degrading enzymes.5
Ligninolytic fungi possess an enzyme system which is a combination of lignin peroxidase (LiP), manganese peroxidase (MnP) and laccase. LiP and MnP are extracellular hemeproteins. LiPs have unusually high redox potential and low optimum pH.6-9 LiP is capable of oxidizing a variety of reducing substrates including polymeric substrates and non-phenolic aromatic compounds.10 MnPs catalyze the oxidation of Mn2+ to Mn3+, which is a strong oxidant that can oxidize phenolic and aromatic amines, yielding organic radicals.11 Laccases are extracellular multicopper oxidoreductases12 that can act on a broad range of substrates, including phenols, phenolic compounds, diamines, aromatic amines, benzenethiol and even some inorganic compounds such as iodine.13 Due to these unusual properties, ligninolytic enzymes are effective in degrading/decolourising synthetic dyes.14-16 White rot fungi coming under basidiomycetes are good producers of ligninolytic enzymes. Most of them are edible and have medicinal properties.
The present study deals with the isolation of a fungal strain producing ligninolytic enzyme followed by the screening for decolorization properties by the selected microorganism.
Collection and isolation of fungal strains
Thirty-two visibly different fungal fruiting bodies were collected from decaying wood and soil litter from various locations of Kerala (Kasaragod, Kangarappady, Ernakulam, Kottayam and Mangalavanam). To obtain pure cultures, small fragments from the inner flesh of the fruiting body were excised and plated onto potato dextrose agar (PDA) (Himedia, Mumbai) plates after surface sterilization with 70% ethanol. Pure cultures were obtained by repeated and successive transfer of mycelia onto new agar plates and were stored at 4 °C on PDA plates.
Screening for production of ligninolytic enzymes
The isolates were grown on lignin containing basal agar medium. The strains growing on this medium were selected for secondary screening, both quantitative and qualitative methods. The qualitative screening was done using PDA plates containing 0.0025% w/v Azure B for detecting lignin peroxidase production,17 and 0.02% guaiacol for detecting laccase production.18
The quantitative screening was done by solid state fermentation (SSF). Pineapple leaves were selected as the substrate for SSF. The leaves were cut to equal size (5x1 cm) and autoclaved at 121 ̊C for 15 min. The moisture content was adjusted to 90% using 0.1 M citrate buffer (pH 5). Three mycelial plugs of size 1 cm2 from 6 days old PDA plate culture were used as inoculum. Inoculated flasks were incubated at room temperature for 5 days.
Extraction of crude enzyme
After incubation, the crude enzyme was extracted by adding 50mL 0.1 M citrate buffer (pH 5) to each flask. The culture supernatant was collected after centrifugation at 10,000 rpm for 10 min at 4 ̊C and stored at 4 ̊C.
Enzyme assays
The activity of LiP, MnP and laccase were checked for each crude extract. LiP activity was measured by the method of H2O2 dependent oxidation of veratryl alcohol to veratraldehyde. The increase in absorbance was checked at 310 nm. MnP activity was determined by the method using phenol red as substrate. The absorbance was checked at 610 nm.19 Laccase activity was measured by the oxidation of ABTS (2,2'-azino-bis(3-ethyl-benzothiazoline-6-sulphonic acid)). The absorbance was checked at 420 nm.20
Enzyme activity was expressed in international units (IU/mL). One IU of enzyme activity was defined as the amount of enzyme that oxidized 1µM of substrate per minute under standard conditions.
Identification of selected strain
DNA was isolated from the strain KSG 32 by the modified method of.21 The isolated DNA was subjected to 18S rDNA amplification using ITS1 and ITS4 primers.22 The reaction mixture contains 25 mM MgCl2, 2 mM dNTP, 1X Taq buffer, 0.5 µM of each primer, 1 unit of Taq polymerase and 100 ng of template DNA. The polymerase chain reaction was performed by initial denaturation for 5 min at 94 ̊C followed by 30 cycles of denaturation at 94 ̊C for 30 sec, annealing at 55 ̊C for 2 min, extension at 72°C for 3 min and a final extension at 72 ̊C for 10 min. The PCR product sequencing was performed on an ABI 3730XL DNA sequencer. The sequence data were submitted to the NCBI database and accession number obtained (GenBank Accession no. KP262029).
The molecular identification of KSG 32 was done by comparing the amplified 18S rDNA sequence with the sequences available in the NCBI nucleotide databases using BLAST (Basic Local Alignment Search Tool) algorithm.23
Dye decolorization experiments
Decolorization studies on various dyes by L. elegans KSG32 were conducted on both PDA and PDB. Initially the ability of L. elegans KSG32 to grow in the presence of various dyes was carried out by growing them on dye incorporated PDA plates. The dyes used were bromophenol blue (SRL, Mumbai), naphthol green B (HiMedia, Mumbai), orange G (HiMedia, Mumbai) and congo red (SRL, Mumbai). The dyes were filter sterilized and added to the sterilized PDA at a final concentration of 0.05 g/L. The dye incorporated plates were inoculated using 0.5 cm2 mycelial plug from 6-day old culture. The plates were incubated at 28 ̊C for 10 days.
In the case of decolourization on PDB, the dyes were added to the medium by either of two methods: a) at the time of inoculation and b) to 5-day old cultures at a final concentration of 0.05 g/L. The flasks were incubated up to 12 days with intermittent shaking at 28 ̊C. At regular intervals, 1 mL samples were withdrawn from the flasks and centrifuged at 10,000 rpm for 10 min. The supernatants were analyzed for the change in absorbance at the absorbance maxima (λmax) of each dye using a UV–visible spectrophotometer (Shimatzu). The λmax of, orange G and congo red is 612 nm, 591 nm, 448 nm and 354 nm respectively.
The percentage of decolorization was calculated in accordance with the following equation:24
Percentage decolorization= (Initial absorbance - Observed absorbance/ initial absorbance) × 100
Toxicity studies of the dyes on fungal growth was monitored during the course of study in terms of biomass content. This was done prior to the decolorization studies in PDB. The toxicity studies were also conducted at a final concentration of 0.05g/L. Biomass was evaluated by determining the dry weight of mycelia. The culture solution was filtered through pre weighed Whatman no. 1 filter paper. The filtered fungal mass was dried at 70 ̊C to a constant weight. The biomass was expressed in g/L.
The enzyme activity during the growth of fungi in control and dye containing PDB on each day of incubation was observed.
FTIR analysis
For Fourier Transform Infrared Spectroscopy (FTIR) analysis, the dye control and decolorized solutions were extracted using an equal volume of ethyl acetate and dried over anhydrous sodium sulphate. The extracts were then dissolved in methanol and used for FTIR analysis (Thermo Nicolet, Avatar 370) in the mid infrared region of 400-4000 cm-1 with a resolution of 4.0 cm-1.
Statistical analysis
All the experiments were performed in triplicates and its statistical significance was checked by performing one-way analysis of variance (ANOVA) with Tukey-Kramer post Hoc test using the SPSS 19 software (SPSS Inc.). Probability values <0.05 were considered to be significant.
Screening for ligninolytic enzymes
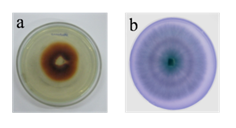
A total of 23 isolates were selected, based on their ability to grow on lignin containing basal media for secondary screening. The oxidative depolymerization of guaiacol forms reddish brown zone by laccase producing strains (Figure 1A), while green colored zones are observed with lignin peroxidase producing strains due to the oxidation of azure B (Figure 1B).
In quantitative screening, these 23 selected isolates were subjected to SSF using pineapple leaves and checked for enzyme production. The enzyme activity for LiP, laccase and MnP in the crude extract are shown in Table 1. After the secondary screening, 9 organisms were found to produce all three ligninolytic enzymes. The production of MnP by most of the isolates was found to be negligible in comparison with LiP and laccase. The strain KSG32 was observed to be having higher laccase (362.7 IU/mL) and LiP (5.17 IU/mL) activity and was thus selected for further studies.
Synthetic Dye |
Mycelial Growth Diameter (cm) |
Decolourization Zone (cm) |
Control |
8.2±0.15 |
- |
Naphthol green B |
8.2±0.1 |
7.6±0.2 |
Bromophenol blue |
7.07±0.11 |
4.7±0.12 |
Orange G |
7.83±0.11 |
4.7±0.1 |
Congo red |
7.67±0.15 |
- |
Table 1 The mycelial growth and decolorization zone on PDA plates 4 days after inoculation of Lenzites elegans KSG32
Identification of the selected strain KSG32
The growth of KSG32 on PDA plate had a regular margin and white color. A woolly mat was formed behind the margins. Concentric rings were also observed (Figure 2A). Hyphal structures and characteristics observed under microscopy showed long skeletal hyphae with irregular branching pattern. Clamp connections were also observed (Figure 2B). The basidiospores were an oblong ellipsoid.
The molecular identification of KSG32 was done by 18S rDNA amplification. Following the BLAST analysis, the isolate was identified to be L. elegans.
Dye decolorization studies on agar plates
During the initial studies on dye incorporated PDA plates, L. elegans KSG32 showed sufficient growth indicating that the fungal growth was not retarded by the dyes. The growth of L. elegans KSG32 on dye containing PDA plates resulted in decolorization zones. A complete decolorization zone was observed in the naphthol green B containing plates. Partial decolorizing zones were observed in orange G, congo red and bromophenol blue containing PDA plates. The growth pattern on the fourth day of inoculation on PDA plates is shown in Figure 3. The mycelial growth diameter and decolorization zone were noted in Table 1.

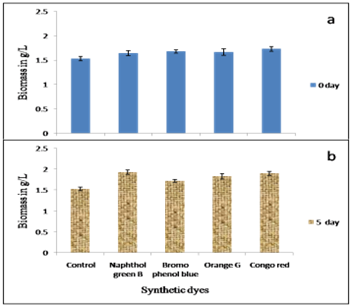
Toxicity studies on fungal growth in terms of biomass content
For the toxicity studies, dyes were added either at the time of inoculation (Figure 4A) or on the fifth day after inoculation (Figure 4B). When the dyes were added at the time of inoculation, the biomass content in the dye containing medium was similar to that of control. Whereas when the dyes were added to the 5-day old culture the biomass content was higher than control, except in bromophenol blue containing media in which the biomass content in the test remains similar to that of the control.
Dye decolorization in liquid culture media
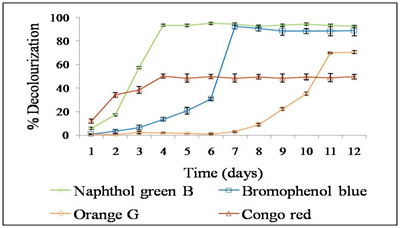
Inorder to quantitatively analyze the ability of L. elegans KSG32 to decolourise dye solutions, the dyes were added to the liquid culture medium. The culture supernatant was analyzed spectro photometrically on daily basis for 12 days. L. elegans KSG32 was capable of decolorizing all the dyes used in the study (Figure 5). In 4 days of incubation 94 % of naphthol green B was decolorized, while 88 % of bromophenol blue decolorization occurred in 7 -day. When incubated for 12 days, 70 % orange G and 48 % of congo red decolorization was observed. Decolorization of orange G starts after 7 days of incubation. Congo red was the least decolorized as only 48 % decolorization occurred. The statistical analysis was done by ANOVA and Tukey-Kramer multiple comparison tests by SPSS software using the mean values of decolorization percentage of all four dyes used in the study. The decolourization percentage of naphthol green B, orange G, congo red were significantly different, while that of naphthol green B and bromophenol blue do not show any significant difference.
As per reports of25 the time at which the dyes were added to the culture medium has an influence not only on the fungal growth but also on dye decolorization. To determine this, the dyes were added to 5-day old culture and the supernatants were collected at 6 h intervals (Figure 6). It is observed that 97% naphthol green B was decolourized in 12 h. Congo red decolorization increased up to 62% when added to 5-day old culture compared to the dye addition along with inoculation. Only 42% orange G decolorization happened when added to the 5-day old culture. Similar decolorization percentage of bromophenol blue showed that the difference in time of addition has no effect on its decolorization, which is similar to the results of toxicity studies. Statistical analysis by comparing means of decolorization percentage of all the dyes showed significant difference.
FTIR spectra analysis
The FTIR spectra of naphthol green B extract from control (a) and after decolorization experiment (b) are shown in Figure 7. The spectrum of the decolourised extract showed significant difference with that of the control.
Enzyme activity during decolorization
Figure 8 shows the significant role of enzyme activity during the growth of the organism in decolorization. There are two aspects for decolorization by fungi, biosorption and enzymatic degradation. In the present study, adsorption of dyes to the mycelium was not observed visually. The presence of laccase and LiP was observed and enzyme activities were monitored daily in control and a dye containing fungal cultures. An increase in laccase activity was observed in the presence of dyes (except for orange G) while LiP activity decreased in the presence of dyes. When the dyes were added to the 5-day old culture, laccase activity increased until the maximum decolorization occurred after that the activity stabilized. This confirms the increased laccase activity in presence of synthetic dyes.
In this study, we found that our isolate L. elegans KSG32, which has been isolated from degrading wood showed higher ligninolytic enzyme production. There are numerous reports on the production of ligninolytic enzymes by basidiomycetes. Several species of white rot basidiomycetes such as Trametes sp., Schizophyllum sp., Pleurotus sp., Ganoderma lucidum produce high amount of these enzymes.18-21 This is the first report on ligninolytic enzyme production by L. elegans. According to22 white rot fungi can be divided into 3 groups based on enzyme production: (i). LiP, MnP and laccase producers, (ii). MnP and laccase producers, (iii). LiP and laccase producers. The most common group is laccase and MnP producers and the rarest group are laccase and LiP producers. L. elegans KSG32 belongs to this group of rare ligninolytic fungi. Some commonly studied white rots and the presence of ligninolytic enzymes are given in Table 2.
White Rot Fungi |
Laccase |
LiP |
MnP |
Reference |
Trametes sp. |
+ |
+ |
+ |
|
Phanerochaete chrysosporium |
+ |
+ |
+ |
|
Pleurotus sp. |
+ |
+ |
+ |
|
Ganoderma lucidum |
+ |
+ |
+ |
|
L. elegans KSG 32 |
+ |
+ |
- |
This study |
Pycnoporus cinnabarinus |
+ |
- |
- |
|
Bjerkandera sp. |
+ |
- |
+ |
Table 2 Commonly studied white rot fungi and their enzyme distribution
Dye decolourization studies
The dye toxicity towards thegrowth of microorganisms is an important factor in decolorization studies. The growth of L. elegans KSG32 on the dye containing PDA plates was not retarded by the presence of the dyes as seen in comparison with the control PDA plate. In the present study the toxicity of dyes was determined by comparing the biomass content of L. elegans KSG32 after 12 -day growth in PDB in the presence and absence (control) of the dye. Studies by26-32 using Phanerochaete chrysosporium, showed a reduction in biomass due to dye toxicity. Increased dye concentrations were found to be affecting the fungal growth.17 However, reports by34 indicate enhanced mycelial growth in the presence of dyes. It is probable that in these cases, the organism may be using the dye as a carbon source. According to,35 an average concentration of 0.3g/L has been reported in the textile effluent. The results indicate that the dyes used for this study were not having any toxic effects on L. elegans KSG32 growth. Instead, of the dyes, it may be acting as an additional carbon source enhancing the fungal mycelial growth.
Bioremediation of synthetic dyes is efficiently achieved by white rot fungi such as P. chrysosporium, due to the low substrate specificity and xenobiotic degradability of its ligninolytic enzymes.7,36,37 In the present study L. elegans KSG32efficiently decolorized naphthol green B which is a metal complex nitroso dye. This adds to the existing reports on theefficient degradation of metal containing dyes using fungi by.38 These dyes are commonly used due to their high light fastness and good wash properties on substrates like wool, nylon and silk.39 The decolorization of naphthol green B under anaerobic conditions by Shewanella oneidensis MR-1 was reported by.40 The commonly used triphenylmethane dye, bromophenol blue decolorization was mainly done by Pleurotus sp.41 According to,42 complex azo dye structures can be completely mineralized by white rot fungi. P. chrysosporium decolorize orange G completely and Pleurotus sajorcaju decolorize it only 50 % was reported by.43
FTIR analysis
Control spectrum showed specific peaks at 3328.80 cm-1 for –OH stretching vibrations, 2966.38 cm-1 and 2866.93 cm-1 for –CH stretching vibrations (Figure 7A). The peak at 2354.62 cm-1 corresponds to the stretching vibrations of cyclic bonds. Peaks at 1657.53 cm-1 and 1383.60 cm-1 assigned to carbonyl stretching vibrations as reported by.44 The peak at 1534.10 cm-1 showed -N=O stretching vibrations.45 Peaks at 1053.54 cm-1 and 1012.94 cm-1 assigned to –SO3Na group vibrations and peak at 687.45 cm-1 for =C-H bending of aromatic rings.46
The spectrum of decolorized extract showed peak shifts and variation of peak intensities when compared with the control dye spectrum. The highly intense peak at 3445.74 cm-1 for –OH stretching vibrations in Figure 7B indicates the increase in oxidation process during the decolorization.13 The disappearance of the peak at 2866.93 cm-1 and the less intense peak at 2974.19 cm-1 showed the decrease in –CH stretching. Also, the intensity of cyclic bond vibrations was decreased when compared with control spectrum. The –N=O stretching vibrations were completely disappeared and the –SO3Na group vibrations were decreased in the decolorized spectrum. There was a shift in =C-H bending from 687.45 cm-1 to 655.31 cm-1 and the peak was widened, this could be due to the decrease in aromaticity. The result shows the involvement of lignin modifying enzymes of white rot fungi in decolorization.
Most studies on dye decolorization focus on laccase due to its ability to degrade both phenolic and non-phenolic compounds. Based on the reports of36 and47 the efficiency of decolorization by laccase increases in the presence of LiP, which helps to overcome the substrate unspecificity of laccase. In the present study, L. elegans KSG32 produce both laccase and LiP which causes more significant decolorization. The results indicate the potential of this organism for future applications in the decolorization and treatment of pollutants in industrial waste water.
L. elegans KSG32 with efficient laccase and lignin peroxidase activity was effectively used for the decolorization of synthetic dyes. The selected dyes were found to be non -toxic towards the organism. This property of the organism can be implemented in the bioremediation of colored industrial waste water.
Authors acknowledge Council of Scientific and Industrial Research (CSIR), India for the financial support in the form of fellowship and Cochin University of Science and Technology for providing laboratory facilities. Authors also acknowledge Sophisticated Test and Instrumentation Centre, Kochi for the FTIR analysis.
The author declares no conflict of interest.

©2017 Lakshmi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.