Journal of
eISSN: 2473-0831


Research Article Volume 11 Issue 2
1Programa de Pós-Graduação em Biotecnologia - PPGBRenorbio, Universidade Estadual do Ceará, Fortaleza, CE, Brasil
2Universidade Estadual do Vale do Acaraú, Centro de Ciências Exatas e Tecnologia, Sobral, CE, Brasil
3Faculdade de Educação, Ciência e Letras de Iguatu, Universidade Estadual do Ceará, Iguatu, Ceará, Brasil
4Universidade Federal do Ceará, Departamento de Análises Clínicas e Toxicológicas, Fortaleza, CE, Brasil
5Universidade Estadual do Ceará, Faculdade de Filosofia Dom Aureliano Matos, Limoeiro do Norte, CE, Brasil
6Fundação Oswaldo Cruz, Laboratório Multiusuário de Pesquisa e Desenvolvimento - Plataforma de Anticorpos e Nanocorpos, Eusébio, CE, Brasil
7Universidade Estadual do Ceará, Centro de Ciências e Tecnologia, Programa de Pós-Graduação Ciências Naturais, Fortaleza, Ceará, Brasil
Correspondence: Hélcio S dos Santos, Universidade Estadual do Ceará, Centro de Ciências e Tecnologia, Programa de Pós Graduação Ciências Naturais, Fortaleza, Ceará, Brasil
Received: February 18, 2022 | Published: March 8, 2022
Citation: Pereira AMG, Bezerra LL, Marinho MM, et al. Inhibition potential of the major secondary metabolites isolated from anacardium occidentale against Mpro enzyme from COVID-19 virus using molecular docking and dynamics studies. J Anal Pharm Res. 2022;11(2):32-37 DOI: 10.15406/japlr.2022.11.00399
The sanitary emergency installed in the world generated by the pandemic COVID-19 instigates the search for scientific strategies to mitigate the damage caused by the disease to different sectors of society. The disease caused by the coronavirus, SARS-CoV-2, reached 216 countries/territories, where about 20 million people were reported with the infection, and more than 740,.000 died. Given the situation, research involving the development and validation of new antiviral molecules, especially against the SARS-CoV-2 virus, is highly relevant. In this work, a in silico study was carried out to evaluate the inhibitory power of phytoconstituents from Anacardium occidentale L. against Mpro enzyme from COVID-19 virus. The molecular docking simulations were performed to understand interactions between some phytochemicals present in A. occidentale against the Mpro enzyme from SARS-CoV-2, the amentoflavone and agathisflavone molecules showed the best affinity energy in the value of -8.0 kcal/mol and -8.3 kcal/mol, respectively. The molecular dynamics results indicated that the agathisflavone ligand had interactions more stable and a highest interaction potential energy with the Mpro enzyme than the amentoflavone ligand.
Graphical abstracts
Keywords: anacardium occidentale, molecular docking, molecular dynamics, SARS-CoV-2
The first reports related to covid (CoV) with severe respiratory syndromes, emerged in China in the early 2000s, posteriorly, in 2002 and 2003, more than 8 thousand cases of the disease and 774 deaths were mentioned, concentrated in eastern and western Asia, as well as reports of deaths in South Africa, Central America and Europe.1 Recently, a new coronavirus was discovered, named SARS-CoV-2, which may cause severe acute respiratory disease in humans.2 This respiratory syndrome formerly known as new coronavirus 2019 (2019-nCoV), is a respiratory disease resulting from infection caused by the new coronavirus, which has enveloped RNAs widely distributed among humans, other mammals, and birds and that cause respiratory, enteric, hepatic, and neurological diseases.3 Besides, the COVID-19 spreads primarily through the respiratory tract, by droplets, respiratory secretions, and contact.4
No specific drug has been approved for coronavirus 2019 disease (COVID-19) to date, as the development of antivirals often takes time. Therefore, evaluation and using of currently available antiviral drugs are essential for a timely response to the current pandemic. In this context, we may highlight the plant species Anacardium occidentale L., popularly known as cashew tree (Figure 1), native to Brazil, found abundantly in the biodiversity of the Northeast, where it stands out for its high socioeconomic importance for the region, representing 99.7% of the Brazilian exports of cashew nuts (the main product of the cashew crop).5 Phytochemical studies report the isolation of several constituents, mainly phenolic compounds (Figure 2). Additionally, this species presents several pharmacological properties such as antimicrobial activity,6 antifungal antioxidant,7 larvicide and antiacetylcholinesterase8 and antidiabetic activity.9 In this work, the inhibition potential of the major secondary metabolites isolated from A. occidentale against Mpro enzyme from COVID-19 virus were evaluate using molecular docking and dynamics studies.
Obtaining the structures
To perform this work, the Pubchem® online molecular repository (http https://pubchem.ncbi.nlm.nih.gov/) was used to obtain the two-dimensional coordinates of the ligands (Agathisflavone (PubChem CID: 5281599), Amentoflavone (PubChem CID: 5281600), quercetin-3-O-rhamnoside (PubChem CID: 5280459), Myricetin (PubChem CID: 5281672), myricetrin-3-O-rhamnoside (PubChem CID: 5281673), gallic acid (PubChem CID: 370), cinnamic acid (PubChem CID: 444539), p-coumaric acid (PubChem CID: 637542), ferulic acid (PubChem CID: 445858), protocatechuic acid (PubChem CID: 72), p-hydroxybenzoic acid (PubChem CID: 135), quercetin 3-O-rutinoside (PubChem CID: 5280805), and Quercetin (PubChem CID: 5280343)), then plotted in the .mol format in the MarvinSketch© software (https://chemaxon.com/products/marvin).10
Structural optimization
For structural optimization, the semi-empirical quantum formalism was used11 through algorithms available in the Molecular Orbital Package code (MOPAC) 2016, Version 16.111W, being used for the optimization method. The Parametric Method 7 (PM7),12 with the Hartree-Fock approximation (HF) (self-consistent field method), for wave function.13 The simulations were configured to perform continuous calculations of cycles of 500 interactions with a convergence value in the order of 10-10 kcal.mol-¹. In this stage, the conformational stability of the compound is given by the total energy, which is the sum of the nuclear repulsive energies with the electronic energy, obtained by solving the12,14 All calculations performed were considering the molecule in the ground state and in a vacuum.
Molecular docking
The structure of the main protease of the COVID-19 virus (Mpro) was obtained in the PDB database, deposited under the code PDB 6M0K, identified as “The crystal structure of COVID-19 main protease in complex with an inhibitor 11b”. As a comparison criterion, it was used the Baricitinib (PubChem CID:44205240), Remdesivir (PubChem CID:121304016), Azithromycin (PubChem CID:447043), and Anakinra drugs (PubChem CID: 139595263) selected in the PubChem repository (https://pubchem.ncbi.nlm.nih.gov/). In preparing enzymatic targets, all residues were removed, and polar hydrogens were added, producing favorable protonation states for molecular docking simulations.15
The molecular docking simulations of the phytochemicals with the COVID-19 were subjected to a hundred independent simulations with cycles of 10 poses each. All simulations were performed using the Lamarckian Genetic Algorithm (LGA) algorithms, implemented in the AutoDock Vina code.16 All simulations were performed with 3-way multithreading and the respective grid box parameters: center_x = -17.744, center_y = 11.691, center_z = 55.282, size_x = 110, size_y = 124, size_z = 116, spacing = 0.575, and exhaustiveness = 8.
The output files (.outligand) generated were rendered in the UCSF Chimera® software,17 where the analysis of interaction and distance of the ligands was made in comparison with the catalytic sites occupied by the control ligand FJC ({N}-[(2~{S})-3-(3-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide). The control ligand was purchased from the original docking deposited at the Protein Data Bank (PDB).
The docking results were clustered into groups with RMSD (Root Mean Square Deviation) less than 2.0 Å.18 To ensure the docking validity, a redocking procedure was conducted using the same parameters for the native ligands from PDB structure.20 The data obtained from the analysis were plotted on the Morpheus® online statistical tool (https://software.broadinstitute.org/morpheus/), where heat maps were generated to identify the ligand-residue interaction profiles (L-R's) and similarity by Pearson's statistical test. The types of L-R's interactions and the Figures were generated using Discovery Studio Visualizer® software.21
Molecular dynamics
The computational simulations with molecular dynamics (MD) were performed through GROMACS (GROningen MAchine for Chemical Simulation) 2020.4 package software22 implemented with the AMBER ff99SB23 force field. First, the best-docked pose of each ligand in the protein was utilized as a starting point for MD simulations. The parameters of Amentoflavone (AME) and Agathisflavone (AGA) molecules were obtained by script ACPYPE (AnteChamber Python Parser InterfacE).24 Subsequently, the solvation system was realized using water molecules described by the TIP3P (Transferable Intramolecular Potential with 3 Points) model,25 and then the system was neutralized with the additional four Na+ ions. The geometry of the system was realized by the steepest descent26 algorithm with 105 steps and an energy tolerance at the value of 10 kJ mol-1 nm-1. Then, each system was equilibrated with 1 ns by ensemble NVT (constant number of particles, volume, and temperature) through of V-rescale thermostat28 at the temperature of 310 K, followed by 1 ns ensemble NPT (constant number of particles, pressure, and temperature) by Parrinello-Rahman29 barostat with a pressure of 1 bar. Finally, the production MD step was performed in 50 ns through Leap-Frog integrator30 with the same temperature and pressure utilized in the equilibrium step. The VMD (Visual Molecular Dynamics)31 software was used to analyze the results obtained by molecular dynamics.
Molecular docking
The molecular docking simulations were performed to understand interactions between some phytochemicals present in Anacardium occidentale against the Mpro enzyme from SARS-CoV-2. Table 1 shows values of affinity energy and the Root Means Square Deviation (RMSD). The remdesivir (REM), anakinra (ANA), amentoflavone (AME), agathisflavone (AGA), ferulic acid (FER), myricetin (MYR), myricetin-3-O-rhamnoside (MOR), quercetin (QUE), quercetin-3-O-rhamnoside (QORh), quercetin-3-O-rutinoside (QORu), azithromycin (AZI), baricitinib (BAR), gallic acid (GAA), p-coumaric acid (PCA), protocatechuic acid (PRA), cinnamic acid (CIA), and p-Hydroxybenzoic acid (PHB) ligands showed values affinity energy of -6.8 kcal/mol, -6.5 kcal/mol, -8.0 kcal/mol, -8.3 kcal/mol, -4.6 kcal/mol, -6.7 kcal/mol, -7.6 kcal/mol, -6.7 kcal/mol, -7.1 kcal/mol, -7.4 kcal/mol, -5.8 kcal/mol, -5.7 kcal/mol, -5.0 kcal/mol, and -4.5 kcal/mol, respectively. All selected poses have RMSD values around 2.000 Å. Therefore, the remdesivir, anakinra, myricetin, myricetin-3-O-rhamnoside, quercetin, quercetin-3-O-rhamnoside, and quercetin-3-O-rutinoside ligands have a good affinity with the Mpro enzyme, due that these molecules had values affinity lower than -6.0 kcal/mol that is the considered standard in the literature.32 In addition, the amentoflavone and agathisflavone molecules showed the best affinity energy in the value of -8.0 kcal/mol and -8.3 kcal/mol, respectively. For this reason, the molecular dynamics simulations were performed only for these ligands.
|
Molecule |
Affinity energy (kcal/mol) |
RMSD (Å) |
|
Agathisflavone |
-8.3 |
1.603 |
|
Amentoflavone |
-8 |
1.513 |
|
Myricetin-3-O-rhamnoside |
-7.6 |
1.759 |
|
Quercetin-3-O-rutinoside |
-7.4 |
1.231 |
|
Quercetin-3-O-rhamnoside |
-7.1 |
1.919 |
|
Remdesivir* |
-6.8 |
1.64 |
|
Myricetin |
-6.7 |
1.703 |
|
Quercetin |
-6.7 |
1.941 |
|
Anakinra* |
-6.5 |
2.058 |
|
Azithromycin* |
-5.8 |
1.253 |
|
Baricitinib* |
-5.7 |
1.379 |
|
Gallic acid |
-5 |
1.471 |
|
Protocatechuic acid |
-4.9 |
1.716 |
|
Ferulic acid |
-4.6 |
1.766 |
|
p-Coumaric acid |
-4.5 |
1.795 |
|
p-Hydroxybenzoic acid |
-4.5 |
1.395 |
|
Cinnamic acid |
-3.7 |
1.692 |
Table 1 Values of affinity energy and RMSD from interactions with Mpro enzyme from SARS-CoV-2 (PDB: 6M0K)
*reference drug
Although the anakinra ligand exhibited a value of RMSD > 2.0 Å, the high affinity indicated interactions stable and probable to occur in biological organisms.18,33 Then, our phytochemicals would be able to interact or inhibit this target stronger, as presented previously.34 The phytochemicals interacted in three sites of interactions in the Mpro enzyme (Figure 3a–c). In the first interaction site, the gallic acid, p-coumaric acid, and protocatechuic acid were localized in the same site that the baricitinib molecule and remdesivir inhibitor (Figure 3a). Figure 3b showed the amentoflavone, quercetin-3-O-rutinoside, agathisflavone, quercetin, quercetin-3-O-rhamnoside, myricetin, ferulic acid, and myricetin-3-O-rhamnoside molecules are localized in the same site that the reference drugs (anakinra and remdesivir). On the other hand, the cinnamic acid and o-hydroxybenzoic acid did not interact in the same regions of reference drugs (Figure 3c).
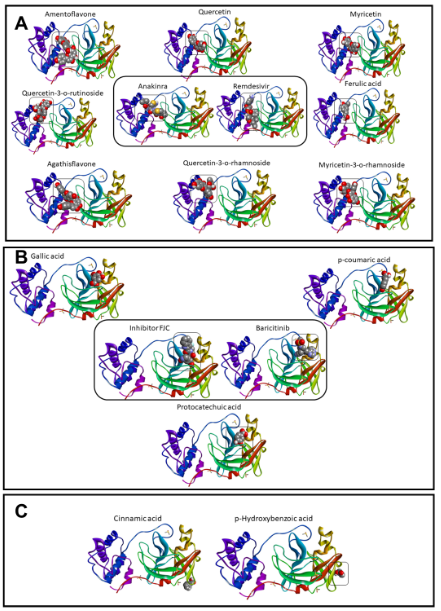
Figure 3 Sites of interactions by the phytochemicals and control molecules with Mpro from COVID-19 virus, which were organized in group of similar interaction with inhibitor FCJ and Baricitinib (A), Anakinra and Remdesivir (B), and molecules without share of interactions (C).
In the group of inhibitor FJC, as presented in Table 2, all molecules shared interaction with the residue of Glu166A, next to residues of His 164A and Met 165A. The gallic acid presented distances of interactions around 2.0 Å with Glu 166A, like FJC, and p-coumaric acid interacted only with this amino acid. The region formed by these residues cited above is essential in the post-transcription process. Besides, the residue of Glu166, and interactions with Ser 144, Gly 143, and Asn142 appear to be related to the binding of inhibitors, being essential to the design of Mpro inhibitors.35,36 Interactions with residues of Gly 143A, Ser 144A, and Cys 145A, close to Asn 142A residue, were also observed. Interactions with Lys 5A, Lys 137A, and Asp 289A were shared between anakinra and remdesivir, as presented in Table 3. However, agathisflavone, ferulic acid, myricetin, quercetin-3-O-rhamnoside, quercetin-3-O-rutinoside, and quercetin performed stronger interactions with Asp 289 than anakinra. Besides, interaction with Glu 288A occurred with remdesivir, agathisflavone, ferulic acid, myricetin-3-O-rhamnoside, myricetin, quercetin-3-O-rhamnoside, and quercetin, but did not occur with anakinra. These same phytochemicals shared with anakinra the interaction with Asp 197A, which did not happen with remdesivir.
|
Residue |
FJC |
GAA |
PCA |
PRA |
|
Interaction distances (Å) |
||||
|
His41 |
- |
- |
- |
- |
|
Gly143 |
2.56 |
2.99 |
- |
2.55 |
|
Ser144 |
3.45 |
2.11 |
- |
2.3 |
|
Cys145 |
1.93 |
2.98 |
- |
2.7 |
|
His164 |
2.36 |
- |
- |
- |
|
Met165 |
3.88 |
- |
- |
- |
|
Glu166 |
1.83 |
1.99 |
3.63 |
3.65 |
|
Pro168 |
- |
- |
- |
- |
|
Gln189 |
3.03 |
- |
- |
- |
Table 2 Hydrogen bonds interactions between FJC group of molecules and Mpro SARS-CoV-2
|
Residue |
REM |
ANA |
AGA |
AME |
FEA |
MOR |
MYR |
QORh |
QORu |
Quercetin |
|
Interaction distances (Ǻ) |
||||||||||
|
Lys5A |
3.23 |
2.67 |
3.22 |
2.67 |
3 |
2.61 |
2.22 |
- |
- |
2.15 |
|
Lys137A |
3.54 |
3.13 |
3.44 |
- |
- |
3.33 |
3.5 |
3.36 |
- |
- |
|
Asp197A |
- |
2.3 |
- |
- |
- |
1.95 |
- |
2.36 |
2.96 |
- |
|
Glu288A |
2.48 |
- |
2.65 |
- |
1.88 |
2.17 |
3.02 |
2.97 |
- |
2.82 |
|
Asp289A |
1.86 |
3.42 |
2.06 |
- |
1.56 |
- |
1.89 |
2.41 |
2.85 |
1.8 |
|
Glu290A |
- |
2.32 |
2.61 |
- |
2.38 |
2.17 |
3.42 |
- |
- |
2.23 |
Table 3 Hydrogen bonds interactions between Anakinra/remdesivir group of molecules and Mpro SARS-CoV-2
Molecular dynamics
The stability of Mpro-AME and Mpro-AGA complexes were analyzed through the Root Mean Square Deviation (RMSD) and the Root Mean Square Fluctuation (RMSF) present in Figures 2a and 2b, respectively. The Mpro-AME complex (Figure 4a) showed the average RMSD in the value of 2.05 Å, and reached equilibrium from 20 ns simulation, while for the Mpro-AGA complex (Figure 4a) was registered average RMSD of 1.65 Å. This system reached equilibrium from 15 ns of simulation. Therefore, the RMSD results indicated that the Agathisflavone ligand had more stable interactions with the Mpro enzyme than the Amentoflavone molecule. The Mpro-AME complex (Figure 4b) showed a higher fluctuation concerning Mpro-AGA. Therefore, the RMSF results indicated that the binding strength of the Amentoflavone molecule with Mpro enzyme was the weakest, while that the Agathisflavone ligand was the stronger. Besides, the Mpro-AGA complex showed good stability because almost all reside of protein were less than 2.0 Å. The Interaction Potential Energy (IPE) of Mpro-AME and Mpro-AGA complexes are present in Figure 5. This energy was obtained by the sum of Coulomb and Lennard-Jones short-range energies in the last simulation frame (50 ns). The Mpro-AME and Mpro-AGA complexes showed the IPE in the value of -129.630 kJ mol-1 and -165.624 kJ mol-1, respectively. Therefore, the agathisflavone ligand showed a higher IPE and consequently a higher affinity with the Mpro enzyme than the Amentoflavone.
The in silico studies represent an initial step for the screening of new potential drugs, being evidenced in this work the potential of the phytochemicals present in the cashew tree against the key enzymes of the replication of SARS-CoV-2, where the biflavonoids amentoflavone and aghatisflavone stand out, these being indicated for future studies in vitro and in vivo. In this perspective, the present work represents a fundamental step for the development of a pharmacological tool to combat SARS-CoV-2, based on the bioprospecting of natural resources present in the Brazilian northeast.
The authors thank the Fundação Cearense de Apoio ao Desenvolvimento Científico e Tecnológico (Funcap), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for financial support and the scholarship. Alexandre M. Rodrigues Teixeira acknowledges the financial support from the CNPq (Grant#: 305719/2018-1). Hélcio Silva dos Santos acknowledges financial support from the PQ-BPI/FUNCAP (Grant#: BP4-0172-00075.01.00/20). Carla F.C. Fernandes thanks the financial support received from project Inova Fiocruz FUNCAP (Grant#:06481104-2020).
The authors declare no conflict of interest.

©2022 Pereira, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.