International Journal of
eISSN: 2574-9889


Case Report Volume 3 Issue 4
1FRIGEs Institute of Human Genetics, FRIGE House, India
2Newcastle University, Australia
3Jena University Hospital, Germany
4NOVA IVI Pulse Womens Hospital, India
5Kanan Maternity and Nursing Home, India
Correspondence: Frenny J Sheth, FRIGE?s Institute of Human Genetics, FRIGE House, Jodhpur Gam Road, Satellite, Ahmedabad-380015, India, Tel 91-79-26921414, Fax 91-79-26921415
Received: November 06, 2017 | Published: December 28, 2017
Citation: Sheth H, Tewari S, Shah K, et al. Prenatal detection of cryptic genomic rearrangement: role of detailed family history, pedigree analysis and advanced genomic technologies. Int J Pregn & Chi Birth. 2017;3(4):295-297. DOI: 10.15406/ipcb.2017.03.00071
Background: Cryptic subtelomeric rearrangements contribute significantly to recurrent miscarriages in couples and are a major cause of undiagnosed non-syndromic dysmorphism and developmental delay in children. Clinical utility of high resolution genomic technologies like subtelomeric FISH and oligonucleotide array aided with precise banding techniques help uncover the causative genetic imbalance and assist in prenatal diagnosis leading to future risk assessment.
Case presentation: We report here a case of five weeks pregnant primigravida married since four years referred for genetic counselling. A family history of developmental delay and dysmorphic features in the proband’s nephew was evident and was inherited from phenotypically normal carrier mother - proband’s sister - who had a cryptic balanced translocation involving #2 and #17 i.e. 46,XX.ish t(2;17)(RP11-321A15-,CTB-50C4+;CTB-50C4-;RP11-321A15+). Normal results from conventional banding of the proband prompted us to perform fluorescence in situ hybridization (FISH), which detected breakpoints at 2q37.3 and in 17q25 region; hence, reassigning karyotype for proband and her sister as 46,XX,t(2;17)(q37.3;q25). Chromosomal microarray (CMA) was carried out at 16weeks of gestation after detection of increased nuchal translucency and single umbilical artery under fetal ultrasonography. Genomic imbalance was evident with 4.9 Mb deletion in 2q37.3 region and 8.2Mb duplication in 17q25.1q25.3 region.
Conclusion: In the genomic era, precise identification of DNA breakpoints may provide useful clues to the underlying genetic anomalies that aid in accurate recurrent risk estimation for a given patient. Careful pedigree analysis and elicitation of family history, together with application of FISH followed by CMA, can detect cryptic imbalance in atypical cases.
Keywords: cryptic subtelomeric translocation, developmental delay, family history, nuchal translucency, subtelomeric fish, chromosomal microarray
ACTG1, actin gamma 1; ARHGDIA, rho-gdp dissociation inhibitor alpha; BDMR, brachydactyly-mental retardation syndrome; CMS, chromosomal microarray; CNV, copy number variant; FISH: Fluorescence in situ hybridization; FSCN2: fascin actin-bundling protein 2, retinal; TWIST2: twist-related protein 2; HDAC4, histone deacetylase 4; GPC1, glypican-1; GPR3, G-protein coupled receptor 3; STK25, serine/threonine kinase 25, NT, nuchal translucency
The association between structural fetal anomalies and chromosome rearrangements has long been established.1 Approximately 5 % of the general population is estimated to be carrier of a balanced rearrangement. Such rearrangements may result in meiotic errors and non-disjunction leading to production of unbalanced gametes. The resulting unbalanced chromosome constitution in gametes may lead to the birth of malformed children, recurrent miscarriage, or infertility.2,3 Most of reciprocal translocations can be detected through conventional cytogenetic techniques. However, carriers of cryptic subtelomeric translocations can be missed by conventional techniques. Current suite of high resolution molecular cytogenetic techniques used in clinical practice include fluorescence in situ hybridization (FISH) that detects targeted anomalies and; oligonucleotide arrays that interrogates the entire genome for copy number variations and sub-microscopic chromosomal anomalies. Detection of these genomic imbalances could inform appropriate genetic counselling and estimate risk of a child in a subsequent pregnancy carrying the imbalance. In the current report, we describe a case where examination of the partial family history together with proband’s medical history informed use of molecular cytogenetic techniques that led to detection of a rare cryptic balanced translocation in the proband.
The proband was a 30-year old female engaged in a non-consanguineous marriage with a 32-years old male. The couple had no previous history of foetal loss. The proband and her husband were referred at 5th week of gestation for genetic counselling to discern their reproductive risk as the nephew of the proband had been diagnosed with dysmorphism and developmental delay. Whilst reports of genetic tests for the affected nephew were not available, FISH report of his phenotypically normal mother i.e. proband’s sister, showed a cryptic balanced translocation involving chromosomes #2 and #17 {46,XX.ish t(2;17)(RP11-321A15-,CTB-50C4+;CTB-50C4-;RP11-321A15+)}. Clinical history of proband’s parents or other family members were not available.
In the first instance, GTG-banding technique at 550 band resolution was performed in the proband according to the standard procedure and chromosomes were classified according to the guidelines by the International System for human Cytogenetic Nomenclature (ISCN, 2013). Proband’s karyotype was found to be normal, however, since the balanced translocation found in the proband’s sister cannot be observed with GTG-banding technique, targeted FISH was carried out using whole chromosome paint probes for #2 and #17 along with BAC clones - RP11-32A15 and CTB-50C-04 for chromosome 2qter and 17qter, respectively. FISH detected breakpoints at 2q37.3 and 17q25 region (Figure 1). Hence, the karyotype for the proband and her sister was reassigned as 46,XX,t(2;17)(q37.3;q25).
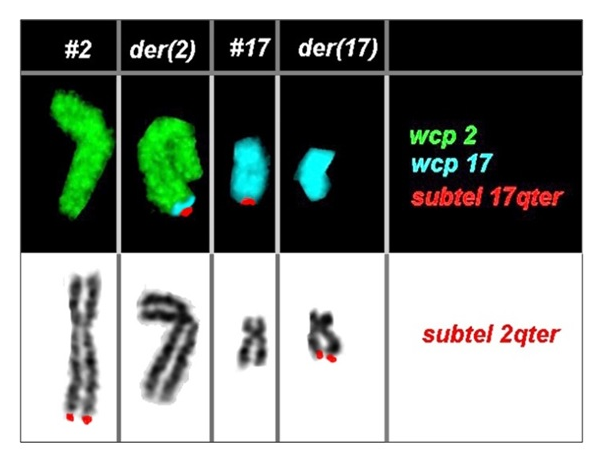
Figure 1 Partial karyotype showing comprehensive characterization of the balanced rearrangement involving chromosome #2 and #17 in the proband using FISH. The proband’s karyotype was identified as 46,XX,t(2;17)(q37.3;q25).
Subsequently, ultrasonography of the proband’s foetus carried out at 12 weeks gestation showed increased nuchal translucency, single umbilical artery and small stomach size. At 16 weeks CMA analysis on the proband’s amniotic sample was performed using Illumina HumanCytoSNP-12 array (Illumina, San Diego, USA) which provided a 30kb resolution. A 4.9 Mb deletion in 2q37.3 region (encompassing 60 genes) and 8.2 Mb duplication in 17q25.1q25.3 region involving 188 genes was detected: 46,XN.arr[hg19] 2q37.3(238093661-243029573)x1, 17q25.1q25.3(72769739-81047565)x3 mat. Proband opted to terminate the pregnancy at the 20th week of gestation in view of the CMA results. Analysis of foetal anatomy showed minor facial dysmorphisms such as round, hypoplastic midface and hypertelorism but no specific signs such as brachydactyly were observed. Foetal autopsy was denied by the parents.
Approximately one in 500 individuals carry a reciprocal translocation. Although balanced translocations are usually associated with normal phenotypes, carriers are at an increased risk of reproductive failure and having offspring with unbalanced rearrangements owing to the abnormal segregation of the rearranged chromosomes.3 Recent developments in the field of molecular cytogenetics have made it possible for submicroscopic rearrangements within chromosomes to be detected using fluorescent in situ hybridization (FISH) and microarray techniques which are not detected by conventional chromosomal banding techniques.4
The current report shows inheritance of derived chromosome #2 along with the normal copy of chromosome #17 from proband to foetus. Genetic imbalance found in the foetus appears to be in congruence with the phenotypes descried in the published literature. Deletion 2q37 syndrome synonymous with Brachydactyly-mental retardation (BDMR) syndrome (OMIM:600430) is caused by chromosomal 2q37 deletions ranging from small sub-microscopic interstitial deletions to large terminal deletions.5,6 Major phenotypes of the BDMR syndrome include mild to moderate developmental delay or intellectual disability, behaviour disorders and brachydactyly type E. Phenotypes may also include facial dysmorphism, congenital heart anomalies, Wilms tumor and urogenital anomalies, epilepsy, eczema, and autistic behaviour.5,6 GPC1, GPR3 and STK25 are the three genes associated with dysmorphism in 2q37 deletion syndrome.7 Recently, haploinsufficiency of histone deacetylase 4 (HDAC4, MIM 605314) has also been identified as the critical genetic mechanism responsible for the major BDMR features in patients with 2q37 deletions. HDAC4 mainly acts as a transcription repressor that alters chromatin structure and influences a broad transcriptional network essential for development and function of brain and connective tissue. TWIST2, another haploinsufficient gene along with HDAC4 may be responsible for the modification of phenotype in 2q37 monosomy. Ablepharon-macrostomia syndrome and Barber-Say syndrome are the two rare ectodermal syndromes which are known to occur due to TWIST2 deletions. The phenotype due to the deletion was difficult to envisage on foetal examination as some facial features are not prominent at 20 weeks of gestation. Several other phenotypes are observed in the literature due to various types of genetic anomalies involving either chromosome #2 only or other chromosomes as well.8 Thus it becomes crucial to report phenotype involving chromosome 17 with 2q37 deletion.
In the present case along with 2q37 deletion, there lies a duplication which has been associated with a well-recognised clinical entity – terminal 17q duplication i.e. 17q25.1q25.3. The anomaly is characterized by psychomotor retardation, microcephaly, skeletal changes, cardiac anomalies and dysmorphic facies.9 Recently, the breakpoints of 17q have been further studied in detail due to the availability of array based technologies. ACTG1, ARHGDIA and FSCN2 in the 17q25 region are the most important genes associated with cardiac malformations.10 The distinct cardiac phenotype is more pronounced in patients with deletion rather than duplications. There were no cardiac malformations observed in our case, thus suggesting that the haploinssuficiency of the genes in 2q37 region might be having a profound effect on the expression of associated phenotypes compared to the protein over-dose due to duplication of the genes in 17q25 region. However, testing this hypothesis through gene expression analysis was beyond the remit of the clinical diagnosis. To the best of our knowledge, this is the first case where simultaneous 2q37 deletion and 17q duplication has been detected prenatally.
Prenatal diagnosis of single umbilical artery with congenital malformations, as in our case, is associated with 25% increased risk of having a chromosomal aneuploidy. This could act as potential marker for the clinician to warrant a thorough foetal assessment using both sonography and genetic testing. Single umbilical artery needs to be assessed as early as 12th week of gestation when an anomalous foetus is suspected in view of the genetic imbalance found in the mother.11 Another important marker detected during early gestational age is nuchal translucency (NT). NT was coined by Nicolaides et al., with 64% sensitivity in detecting trisomy 21.12 However, the use of molecular karyotyping in increased NT with normal foetal karyotype still has a considerable lacuna. Recently, Grande at al.13 systematically reviewed and showed that genomic microarray provides a 5% incremental yield of detecting pathogenic copy number variants (CNV).13 In our case, we were aware of the higher likelihood of detecting a CNV due to a maternal balanced translocation and nuchal translucency that has proved to be a sensitive marker for genomic imbalances. Single umbilical artery and increased NT are sensitive ultrasound markers and need high resolution diagnostic modalities when foetal karyotype reports appear normal.
We would like to state that application of high resolution genomic approaches has largely overcome the limits of banding cytogenetics in diagnosing sub-microscopic genetic aberrations in patients with “normal” karyotype. Nevertheless, critical evaluation of pedigree and reproductive as well as family history can provide valuable clues to formulate a clinical hypothesis that can later be tested using the latest genomic techniques, as was evident in the current report by predicting the recurrence risk for the couple and offering prenatal/preimplantation diagnosis for future conceptions.
We thank the patient for cooperation. Authors thank Dr. Sunil Trivedi for critical reading and useful suggestions.
The authors declare that they have no financial or non-financial competing interests.
Informed written consent was obtained from all the participants for publication of their clinical details and/or clinical images. A copy of the written consent is available for review by the journal editor.

©2017 Sheth, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.