International Journal of
eISSN: 2470-9980


Research Article Volume 2 Issue 6
1Department of Veterinary Microbiology and Parasitology, University of Maiduguri, Nigeria
2Department of Veterinary Pathology and Microbiology, Ahmadu Bello University, Zaria, Nigeria
Correspondence: Musa JA, Department of Veterinary Microbiology and Parasitology, University of Maiduguri, Nigeria, Tel +234 8030575355
Received: August 07, 2016 | Published: October 18, 2016
Citation: Musa JA, Kazeem HM, Raji MA, Useh NM (2016) Prevalence and serological detection of Enterohaemorrhagic Escherichia coli O157 serogroup in commercial cattle farms in Kaduna State, Nigeria. Int J Vaccines Vaccin 2(6): 00053. DOI: 10.15406/ijvv.2016.02.00053
Objectives: To determine the prevalence and to detect the presence of Escherichia coli O157 serogroup in commercial cattle farms in Kaduna State, Nigeria.
Methods: A total of 240 faecal samples were obtained from eight randomly selected commercial cattle farms and then placed in tryptose soya broth (TSB). Thereafter, the faeces were inoculated onto sorbitol MacConkey agar (SMAC) to identify non-sorbitol fermenting (NSF) colonies, and then sub cultured onto eosin methylene blue agar (EMB). Escherichia coli O157 agglutination test for the identification of E. coli O157 antigen was carried out with O157 latex kit (Oxoid). This involved mixing the isolates with 2ml of 0.85% saline solution separately, followed by the addition of test antigens to observe for agglutination.
Results: Colonies on SMAC appeared phenotypically colourless and were presumptive for E. coli O157, while those on EMB gave the characteristic greenish metallic sheen for E. coli. Of the 240 faecal samples, E. coli colonies of seventy six (31.2%) were confirmed by Gram staining and biochemical testing using Indole, Methyl red, Voges Proskauer and Citrate (IMViC). Characterization of the E. coli isolates detected two O157 serogroups from two apparently healthy cattle. The prevalence of E. coli O157 was found to be 0.8%. Association between the serogroup and source of samples (farms) was significant (P<0.05). The study confirmed that cattle are important source of enterohaemorrhagic E. coli and may pose a risk to humans who come in contact with cattle faeces in areas of Kaduna State, Nigeria.
Conclusion: Escherichia coli O157 serve as a threat to human health. The differences in the dynamics of disease may contribute to disparity in prevalences. Good hygienic measures on the farms are essential in limiting the transmission of E. coli to in-contact individuals.
Keywords: detection, enterohaemorrhagic, e. coli o157, serogroup, prevalence, cattle, Kaduna, Nigeria
The term ‘Enterohaemorrhagic Escherichia coli’ (EHEC) was originally used to describe strains that cause haemorrhagic colitis (HC) and haemolytic-uraemic syndrome (HUS), express shiga-toxins (Stx), cause attaching and effacing (A/E) lesions on epithelial cells and possess large plasmid.1 On the other hand, they are known as Shiga toxin-producing Escherichia coli (STEC); foodborne pathogens that are associated with human illnesses that could be life-threatening in nature.2 They have emerged through the production of shiga toxins; stx1 and stx2 and other probable virulence factors such as intimin gene (eae) and enterohaemolysin (ehly).3‒4
The major and the most essentially suggested cardinal feature of EHEC strains is the production of shiga toxins (stx1 and stx2), which comprise a family of structurally related cytotoxins with similar biological activity and distinct antigenic structures.1 The colonization of the intestinal mucosa by most of the EHEC is associated with a mechanism that subverts the function of the epithelial cells. The effect of this interaction is the inducement of a characteristic “attaching and effacing” (A/E) lesion, a complex mechanism genetically controlled by a locus of large pathogenicity island (PAI) called the “locus of enterocyte effacement (LEE)”. Intimin mediates the intimate attachment of EHEC by binding to β1-integrins and to cell-surface localized nucleolin.5
The EHEC can cause diarrhoea, HC and HUS in children less than five years of age, the elderly and the immunocompromised individuals.6,7 Escherichia coli (E. coli) O157 was first associated with HC in humans in 1982 and was earlier isolated and reported in cattle with substantial evidence provided by serological studies that, E. coli O157:H7 is widely spread in cattle.8,9
Escherichia coli O157 in human infections may either be waterborne, food borne or consumption of food and water contaminated by faeces of ruminants, direct contact with infected animals or human-to-human transmission.10 In EU member states, data on STEC in cattle and beef products are poor. There has been low isolation rate for positive samples were 0.1 per cent for STEC O157 and low levels of microorganisms.11
Enterohaemorrhagic E. coli is a diverse group of food borne zoonotic pathogens of which O157:H7 is a major public health concern and is reported as an emerging infectious agent.12,13 Nearly 75 per cent of human STEC O157 outbreaks in the USA originated from food of bovine origin, especially the undercooked ground beef.14 In EU member states, except for Portugal, a notification rate of 1.15 cases/100,000 population was recorded in 2012, for a total of 5671 confirmed cases, with increasing trends during 2008–2012 observed in different countries, including Italy. In Europe, serogroups O157 was one of the most prevalent, representing 56 per cent of the STEC isolates. STEC O157 was isolated from 33 per cent of cases in Italy.11
Cattle appear to be the main reservoir of EHEC and the organism has also been isolated from healthy animals.15,16 Since early 1990s there has been a global increase in EHEC infections. Ever since, EHEC have been of considerable concern not only because of severity of the illness they can cause, but also the low infectious dose and increasing incidence worldwide. Currently about 450 O:H serotypes of STEC have occurred.17 Sorbitol MacConkey (SMAC) agar is used for the presumptive identification of E. coli O157, while E. coli is most often confirmed from faeces on eosin methylene blue (EMB) agar.18
Study design
The study was a cross-sectional study (prevalence study): an observational study that involves the selection of a sample of individuals from a larger population, and then the determination, for each individual of the simultaneous presence or absence of disease or causative agent(s) of a disease and hypothesized risk factor.
Sample size
The sample size was determined according to the method of.19
Study area
The study area is Kaduna State, which is located between latitude 100 and 110N and longitude 7o and 8oE in North-central part of Nigeria (Figure 1).
Sample collection
A total of two hundred and forty (240) faecal samples from two hundred and thirty three (233) apparently healthy and seven diarrhoeic cattle were collected from eight randomly selected commercial farms in Kaduna State using stratified random sampling technique. The farms were designated as farms A (FA), B (FB), C (FC), D (FD), E (FE), F (FF), G (FG) and H (FH). All the farms were located in five different Local Government Areas of Kaduna State (see Fig.1). Animal features like age (young: 0-1year, and adult: >1year ≤ 2years, >2years ≤ 3years and >3years), sex (male and female) and breeds (exotic: Friesian, Holstein and Simmental, and local or indigenous: Bunaji, Rahaji, Sokoto Gudali, and Adamawa Gudali) were obtained using farm records. Faecal material of 1-2g was aseptically collected from the rectum of each animal using clean disposable hand gloves. The samples were placed in separate sterile bottles containing 8-9ml of tryptone soya broth (TSB), kept in a cool box containing ice blocks, and then transported to the Bacteriology Laboratory, Department of Veterinary Pathology and Microbiology, Faculty of Veterinary Medicine, Ahmadu Bello University, Zaria, where they were processed immediately.
Isolation and identification of suspected colonies
The bacterial isolation and identification were carried out by standard procedures described previously.20,21 The cattle faecal samples from tryptone soya broth (TSB, CM0129; Oxoid, Basingstoke, UK) were inoculated onto sorbitol MacConkey agar (SMAC, CMS0813 Oxoid, Basingstoke, UK) and then incubated for 24-48hours at 370C. The non-sorbitol fermenting (NSF) colonies on SMAC agar were identified and then sub cultured onto eosin methylene blue agar (EMB, VM62604262; Merck, USA) and the plates were incubated at 37oC for 24- 48hours. Positive E. coli isolates on EMB were identified and then subjected to biochemical tests.
Biochemical characterization
Colonies of E. coli growing on eosin methylene blue agar plates were subjected to biochemical tests: Indole, methyl red, Voges-Proskauer, citrate (IMViC), motility and triple sugar iron (TSI) test.
Serogrouping of somatic O isolates
All confirmed E. coli isolates were sub-cultured onto nutrient agar slants and stored at 40C for serogrouping.22,23 Escherichia coli O157 agglutination test for the identification of E. coli O157 antigen was carried out with O157 latex kit (Oxoid). This involved mixing the isolates with 2ml of 0.85% saline solution separately, followed by the addition of test antigens to observe for agglutination.
Statistical analysis
The results were analyzed using Chi-square two by two contingency table with Statistical Package for Social Sciences (SPSS) 14.0 version and Microsoft Excel version 2010. The value of P<0.05 indicated statistical significance (Table 1)24.
Farms |
Location of the Farms In their Respective L.G.A |
Positive E. coli O157 [n (%)] |
Farm A (FA) |
Giwa |
0 (0.0) |
Farm B (FB) |
Chikun |
1 (3.0) |
Farm C (FC) |
Igabi |
1 (3.0) |
Farm D (FD) |
Kachia |
0 (0.0) |
Farm E (FE) |
Igabi |
0 (0.0) |
Farm F (FF) |
Igabi |
0 (0.0) |
Farm G (FG) |
Igabi |
0 (0.0) |
Farm H (FH) |
Sabon Gari |
0 (0.0) |
Total |
|
2 (0.8) |
Table 1 Distribution of E. coli O157 serogroup among commercial cattle farms in Kaduna State, Nigeria
P < 0.04
The characteristic morphological appearance presumptive of E. coli O157 on sorbitol MacConkey agar was found to present non-sorbitol fermenting colourless colonies (Plate I), while on EMB E. coli exhibited greenish metallic sheen as shown on Plate II. The age distribution of E. coli O157 serogroup isolated from commercial cattle farms has shown that two (6.7%) O157 serogroups were isolated from adults within the age group of greater than 2years but less than or equal to 3years. The relationship observed between age and E. coli serogroup from commercial cattle farms was not statistically significant (P>0.05) (Table 2).
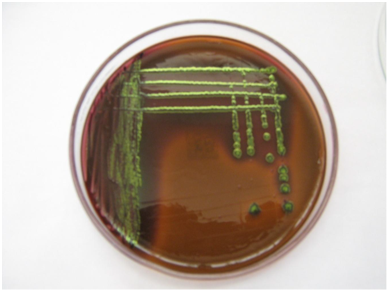
Plate 2 Colonial appearance of E. coli on eosin methylene blue (EMB) agar showing characteristic greenish metallic sheen.
Age |
|
Specific Prevalence Rate (%) |
Positive E. coli O157 (%) |
Young |
0-1yr |
0 |
0 (0.0) |
Adult |
>1 ≤ 2years |
0 |
0 (0.0) |
>2 ≤ 3years |
6.7 |
2 (6.7) |
|
>3years |
0 |
0 (0.0) |
|
Total |
2.8 |
2.8 |
2 (0.8) |
Table 2 Age distribution of E. coli O157 serogroup detected in commercial cattle farms in Kaduna State, Nigeria
Young = 0-1 year, Adults = >1≤ 2 years, >2 ≤ 3 years and >3 years
The relationship between breeds of cattle and E. coli serogroup from commercial farms was analyzed. The specific prevalence rate ranges from 0.0% in exotic breed (Friesian, Holstein and Simmental) to 3.3% in local breed (Rahaji and Bunaji). However, O157 was found only among Rahaji and Bunaji breeds but absent in others (Table 3).
Breeds |
|
Specific Prevalence Rate (%) |
E. coliserogroup O157 |
Exotic |
Friesians |
0 |
0 (0.0) |
Holstein |
0 |
0 (0.0) |
|
Simmental |
0 |
0 (0.0) |
|
Local (Indigenous breeds) |
Bunaji |
3.3 |
1 (3.3) |
Rahaji |
3.3 |
1 (3.3) |
|
Sokoto Gudali |
0 |
0 (0.0) |
|
Adamawa Gudali |
0 |
0 (0.0) |
|
Total |
|
2.8 |
2 (0.8) |
Table 3 AmplitudeRelationship between breeds and E. coli O157 detected incommercial cattle farms in Kaduna
state, Nigeria
Exotic= Friesians, Holstein, Simmentals with their crosses Local (Indigenous breeds) = Bunaji (White Fulani), Rahaji, Sokoto Gudali and Adamawa Gudali).
The relationship between sex and E. coli serogroup isolated from commercial cattle farms indicated that E. coli serogroup was distributed according to the sex of animals. Zero percent (0.0%) of O157 serogroup were found in males and 2 (1.0%) in females. The overall prevalence is found to be 0.4% for both male and female cattle (Figure 2).
Escherichia coli O157 serogroups expressed a characteristic sorbitol non-fermenting phenotype used in many studies for their identification in cultures of faecal and food samples. This concurs with the earlier report.25 The distribution of the E. coli O157 serogroup and the sources of samples (commercial cattle farms) were investigated. The specific prevalence of E. coli O157 was 3.0% each from FB and FC. A prevalence of 0.8% for E. coli O157 was found to be statistically significant (P<0.05). This is in consonance with the previous findings, who reported a prevalence of 0.7% in apparently healthy cattle in Spain.23
Although higher prevalence of up to 7% was reported in apparently healthy cattle from the South-western part of Nigeria,26 4.5% in apparently healthy cattle in Borno and Adamawa States,27 2 % from apparently healthy cattle in Lagos,28 and 51.4 % from the faeces of cattle in England and Wales.29 These prevalences appeared to be at variance with the findings of this study. Although, studies in Nigeria confirmed that E. coli O157 is present in commercial cattle farms in Kaduna State. The variation in prevalence may possibly be attributed to variation between areas of study and the dynamics of disease. However, the relatedness of these factors to the presence of E. coli O157 in cattle remained to be extensively determined.
In this study, the sex distribution of E. coli O157 was found to occur. The males presented 0.0% prevalence of E. coli O157 serogroup as compared to their male counterpart that had 1.0% prevalence. The overall prevalence is found to be 0.4% for both male and female cattle sampled. Although, previous findings reported 11.6 % and 3.0 % isolation rate in cows and bulls using DNA hybridization technique, they worked on serogroups O157 and others that were not reported in this study. Consequently, differences in techniques might serve as contributory elements to these disparities observed.30
The significance of E. coli O157 serogroup as a food-borne pathogen contributes greatly to the relevance of this study. It is recommended that contact with cattle faeces, particularly during milking and meat processing, sanitation on the commercial farms and strict hygienic measures should be inculcated in daily practices to serve as preventive measures from been infected by E. coli O157. In conclusion, the investigation revealed that one serogroup of E. coli O157 each was found in FB and FC. Escherichia coli O157 serogroup were harboured by cattle in commercial farms. Therefore, the presence of EHEC in food is likely to occur through contamination with cattle faeces. This further emphasizes that cattle are important reservoir of enterohaemorrhagic E. coli O157 which may pose a risk to humans that come in contact with cattle faeces.
We sincerely acknowledge Mr Michael Udele, Dodo Bawa and Salamatu Garba of Bacteriology Diagnostic Laboratory, Department of Veterinary Pathology and Microbiology, Ahmadu Bello University, Zaria, for valuable contributions.
Author declares there are no conflicts of interest.
None.

©2016 Musa, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.