International Journal of
eISSN: 2573-2889


Review Article Volume 4 Issue 1
Donetsk Physical and Technical Institute NAS of Ukraine, Ukraine
Correspondence: Helen A Grebneva, Donetsk Physical and Technical Institute NAS of Ukraine, Kiev, Ukraine
Received: December 17, 2018 | Published: January 10, 2019
Citation: Grebneva HA. Paradigm change in mutagenesis: polymerase tautomeric models for targeted, delayed and untargeted ultraviolet mutagenesis during error prone and SOS replication of double stranded DNA, containing cis-syn cyclobutane thymine dimers or thymine in rare tautomeric forms. Int J Mol Biol Open Access. 2019;4(1):1?15. DOI: 10.15406/ijmboa.2019.04.00091
Polymerase-tautomeric models for targeted ultraviolet mutagenesis is developed that are based on the formation of rare tautomeric bases in DNA bases. Five rare tautomeric forms may form for thymine and adenine. These rare tautomeric forms will be stable if corresponding nucleotides are part of cyclobutane pyrimidine dimers or are in small neighbor of the cyclobutane dimers and during DNA synthesis. It is shown that during error-prone or SOS synthesis the modified or specialized DNA polymerases insert canonical bases opposite the cis-syn pyrimidine cyclobutane dimers or DNA bases in rare tautomeric forms; the inserted bases are capable of forming hydrogen bonds with bases in the template DNA. Structural analysis indicates that one type of cis-syn cyclobutane thymine dimers containing a single tautomeric base (TT*1, with the ‘*’ indicating a rare tautomeric base and the subscript referring to the particular conformation) can cause A:T®G:C transition or homologous A:T®T:A transversion. The cis-syn cyclobutane thymine dimers containing T*4 result in A:T®C:G transversion, while TT*5 dimers can cause A:T®C:G transversion or homologous A:T®T:A transversion. Structural analysis indicates that opposite cis-syn cyclobutane thymine dimers TT2* it is impossible to insert any canonical DNA bases with the template bases with hydrogen bonds formation. Cis-syn cyclobutane thymine dimers wherein a thymine is in the rare tautomeric form T2* may result in targeted insertions and targeted deletions. Delayed mutations are an important part of radiation-induced genomic instability. Structural analysis of the insertion of the bases showed that opposite rare tautomeric form of thymine T3* adenine can be incorporated, but may be inserted any other canonical base so that between them hydrogen bonds are formed. Opposite canonical thymine cytosine can be incorporated only. If in the synthesis of DNA containing the cis-syn cyclobutane dimers TT3*, involved DNA polymerases with relatively high fidelity of synthesis, mutations not appear. However, if further DNA synthesis will involve DNA polymerases having a low fidelity of synthesis, there may be base substitution mutations after DNA has been damaged. Canonical cis-syn cyclobutane thymine dimers TT may result in targeted delayed transversions T-A®G-C only, cis-syn cyclobutane thymine dimers TT3* may result in targeted delayed transitions T-A®C-G, targeted delayed transversions T-A®G-C and T-A®A-T. Currently, untargeted mutations are studied in the context of radiation-induced bystander effects. Untargeted base substitution mutations are base substitution mutations then one or some nucleotides are inserted in DNA molecule on, so called, undamaged sites of DNA. Thymine in rare tautomeric forms T1*, T4* and T5* may result in untargeted base substitution mutations. Thymine in rare tautomeric forms T2* may result in untargeted insertions.
Keywords: UV mutagenesis, radiation induced bystander effects, radiation induced genomic instability, rare tautomeric forms, targeted base substitution mutations, targeted insertions, targeted deletions, untargeted base substitution mutations, untargeted insertions, targeted delayed base substitution mutations, cis-syn thymine cyclobutane dimers, error prone replication, SOS replication
Some features of the ultraviolet mutagenesis
Ultraviolet radiation produces cyclobutane pyrimidine dimers under induced by UVB irradiation.1,2 Cis-syn cyclobutane pyrimidine dimers are a large majority of mutations induced by ultraviolet light.3,4 In cells of E. coli, irradiation with a wavelength of 260 nm produces about 40% of thymine dimers, 5%-19% of cytosine dimers and 19%-22% of dimers consisting of cytosine and thymine. For UVC and UVB, the total relative proportion of cyclobutane pyrimidine dimers formed in the thymine-thymine, thymine-cytosine, cytosine-thymine and cytosine-cytosine sites was about 28%, 26%, 16% and 30%, respectively. However, for UVA, cyclobutane pyrimidine dimers were formed much more frequently in the thymine-thymine sites than at the thymine-cytosine, cytosine-thymine and cytosine-cytosine sites (57% vs. 18, 11 and 14%, respectively).5 Cyclobutane pyrimidine dimers are effectively removed by excision repair.6 If not all dimers are moved cis-syn cyclobutane thymine dimers may produce mutations.7 Mutations occur during error-prone and SOS synthesis.8–11 They cause targeted base substitution mutations12,13 targeted insertions targeted deletions, targeted complex mutations and targeted delayed mutations.14–19 Only 5-12 % of cis-syn cyclobutane pyrimidine dimers result in mutations.20 Mutations occur opposite the cyclobutane pyrimidine dimers is termed targeted mutations.8–11,20,21 Mutations are formed in the vicinity of the damage are termed untargeted mutations.22 Long-wave ultraviolet UVA light can cause delayed mutations.23,24 The delayed mutations are usually point mutations, more than half of them are base substitution mutations. As the experiment shows, DNA damage leading to delayed mutations is usually not removed. Delayed mutations can make a significant contribution to genetic diseases.25,26
At present, the conventional paradigm relates the reason of mutations exclusively to sporadic errors of DNA polymerases. It is assumed that the mutations arise because the DNA-polymerase sometimes incorporates non complementary nucleotides opposite the cyclobutane pyrimidine dimers.27–29 In the articles,30–32 the authors suggested that mutations induced by ultraviolet light occur only after deamination of the cytosine or 5-methylcytosine within the pyrimidine dimer. Oxidative damage of the bases is considered the cause of mutagenesis under the action of long-wave ultraviolet light (UVA).29 According to33 7,8-dihydro-8-oxoguanine makes an important contribution to the genotoxicity of UVA irradiation of the yeast Saccharomyces cerevisiae. Watson and Crick34 suggested that spontaneous mutations are formed when hydrogen atoms are attached to the bases of DNA, or bases lose hydrogen atoms due to contact with water molecules; which influences the character of base pairing. The participation of rare tautomeric forms in mutagenesis was repeatedly discussed.35–38 A large number of works devoted to the study of rare tautomeric forms have been performed, both in DNA bases and in model molecules.39 It was shown that after cytosine was irradiated with UV light (cytosine was isolated in a low-temperature argon matrix), it changed from the main tautomeric form to rare tautomeric forms, their ratio depended on the intensity of irradiation.40 In Ref39 the nature of the defect states in crystals of bases of nucleic acids irradiated with UV light by the method of thermally stimulated luminescence was studied. It was concluded that there are rare tautomeric forms of cytosine in the investigated crystals39 However, all currently existing mutagenesis models cannot explain most of the phenomena of mutagenesis.41,42 Polymerase paradigm.27–29 tautomer model by Watson and Crick34–37 and deamination model30–32 claim an explanation targeted base substitution mutations only. Cyclobutane pyrimidine dimer consisting deamination cytosine or 5-methylcytosine30–43 may result in base substitution mutations. Some data implicate the deamination of cytosine to uracil as a possible cause, but other results appear to indicate that the rate of deamination is too low (in the range of 10-10sec-1 by in vitro measurement) for this to be significant in Escherichia coli.44 The experimental data on the incorporation of DNA bases by various DNA polymerases were summarized and called A-rule.28 It turned out that opposite the cis-syn cyclobutane thymine dimers DNA polymerase inserted adenine most often44 but sometimes they inserted guanine, thymine or cytosine.45–48 In the polymerase paradigm, it is assumed that both matrix and inserted bases are in canonical tautomeric forms. We make a structural analysis of the inserting of the canonical bases found in the study of A-rule in the papers opposite cis-syn cyclobutane thymine dimers or (6-4) adducts.49–50 The canonical tautomer of guanine (Figure 1) and the canonical tautomer of thymine (Figure 1c) cannot form hydrogen bonds with canonical tautomers of thymine for steric reasons. But canonical tautomeric forms of cytosine can be incorporated opposite the canonical tautomer of thymine (Figure 1b). Specialized and modified DNA polymerases incorporates canonical bases capable of forming hydrogen bonds with cis-syn cyclobutane pyrimidine dimers in template DNA.51–62 Therefore, the polymerase paradigm cannot explain the mechanism for the formation of targeted base substitution mutations (for a more detailed analysis see in50 In my opinion, now mutagenesis as an area of research is in deep crisis.

Figure 1 Possible formation of pairs between: a) the canonical tautomer of thymine and the canonical tautomer of adenine; b) the canonical tautomer of thymine and the canonical tautomer of cytosine; c) the canonical tautomer of thymine and the canonical tautomer of guanine; d) the canonical tautomer of thymine and the canonical tautomer of thymine.49
The idea of Watson and Crick that rare tautomeric forms of DNA bases can play an important role in mutagenesis34 is certainly magnificent, but requires further development. To understand the mechanisms of the formation of different mutations under the action of different mutagens, it is necessary to understand what happens when at least one mutagen acts. As such a model, in my opinion, ultraviolet mutagenesis is best suited. I believe that in order to understand how mutations are formed under the action of ultraviolet light, the following should be done. It is necessary to study the processes that occur when an ultraviolet quantum of energy interacts with a DNA molecule. It is necessary to see to what chemical changes of DNA structure this can lead. It is necessary to study the conditions under which these chemical changes will be stable. It is necessary to study what mutations they can lead to in the case of error prone or SOS replication of DNA that has such damage. This plan was implemented in several article cycles.63–87 A semiempirical potential function capable of describing hydrogen bonds with lengths different from equilibrium has been developed63,64 It was used to find potential curves of the guanine-cytosine pair for several lengths of hydrogen bonds.63,65 The obtained curves were used to study the nature of the vibrations of atoms and atomic groups for isolated bases and guanine-cytosine base pair for hydrogen bonds in the ground and excited states.63,65,66 The problem was solved for an isolated guanine-cytosine pair63,65 and a pair of guanine-cytosine located in the DNA strand.63,66 The theory of heat deexcitation of hydrogen bond protons in paired bases of DNA molecules was developed.63,67 The previously obtained results63,67 made it possible to estimate the lifetime of the excited hydrogen bond with respect to thermal transitions. The processes of propagation of excitation energy along the DNA molecule were studied, a new quasiparticle, a proton exciton, was predicted, and its properties were studied.68 It turned out that the main contribution to the process of hot and cold spots of ultraviolet mutagenesis formation is made by the processes of propagation of excitation energy along the DNA molecule.69 All these results were used in the construction of a model for rare tautomeric forms of DNA bases formation upon irradiation of a DNA molecule with ultraviolet light.63,75 The rare tautomeric forms are stable when the respective bases are involved in cyclobutane thymine dimers. This is because the DNA strand bends once pyrimidine dimers arise, and the hydrogen bonds between the bases are broken between the bases that neighbor the cyclobutane pyrimidine dimers.88,89 The rare tautomeric forms of DNA bases are stable in DNA synthesis.76 These conclusions were confirmed by experiments.61,62 The results of studies on the structure of the active centers polymerases show that the bases in rare tautomeric forms may exist in the active sites of DNA polymerases.61,62 These results served as the basis for the development of the polymerase-tautomeric models for targeted ultraviolet mutagenesis,41,42,50,63,69,70,72,73,75,76,78–82 radiation-induced bystander effects71,74,77,84,85 and radiation-induced genomic instability.49,63,83,86 The polymerase-tautomeric models are based on Watson and Crick’s hypothesis that the mutagenesis is based on the ability of the bases to change the tautomeric state. I propose the mechanisms of targeted base substitution mutations formation during error-prone or SOS synthesis of DNA containing cis-syn cyclobutane cytosine and thymine dimers.41,42,75 I propose the mechanisms of targeted insertions formation during error-prone or SOS synthesis of DNA containing cis-syn cyclobutane cytosine78 and thymine dimers.42,79 A mechanisms was proposed for targeted complex insertions42,81 and targeted deletions42,80 caused by cis-syn cyclobutane thymine dimers.
The mechanism of rare tautomeric forms formation in DNA base pairs
A mechanism for changes in the tautomeric state of base pairs has been propose.90 The destiny of DNA-absorbed UV-quantum significantly depends on several factors. On the one hand, it depends on nucleotide composition of the neighboring pairs of bases and, on the other hand, on the relation between the lowest singlet (short-lived) and triplet (long-lived) levels of energy of various bases.75 It has been shown, that the tautomeric changes can occur at no radiative de excitation of the DNA, which has absorbed the UV-quantum from triplet levels of energy owing to strong forced oscillations. Such oscillations result in changes of lengths of hydrogen bonds. The hydrogen bonds that are formed between the DNA bases are characterized by a strong valence bond with one of the partner atoms in the Н-bond, and a weak bond with the other. When the Н-bond length changes, the length of a valence bond changes very little. The distance from the hydrogen to the second atom, however, varies considerably. When the hydrogen bond becomes shorter, atom of hydrogen is almost in the center of hydrogen bond. When the Н-bond is extended, the hydrogen atom can assume new position.
It was assumed that the tautomeric state of the constituent bases may change during the formation of cyclobutane pyrimidine dimers.75 A mechanism for changes in the tautomeric state of base pairs has been proposed for the case when DNA is UV-irradiated and cyclobutane pyrimidine dimers are formed.75 The rare tautomeric forms of bases are stable at cis-syn cyclobutane pyrimidine dimers formation and in DNA synthesis.90 The forms are stable when the respective bases are involved in cyclobutane thymine dimers.75 The rare tautomeric forms of bases are stable because, at cis-syn cyclobutane pyrimidine dimers formation, the DNA strand is bent and the hydrogen bonds between the bases are significantly weakened or are broken.88,89 When the hydrogen bond becomes weaker it becomes longer. As shown in91 in this case, there is a second minimum. To find out what new tautomeric forms of DNA bases can be formed, I used the structural - dynamic model of semi-open states of the DNA by Hovorun.92 Five new rare tautomeric conformations of adenine and thymine base pairs (Figure 2)75 and seven new rare tautomeric conformations of G:C base pairs are proposed that are capable of influencing the character of base pairing.41 It is well known that the A:T base pair contains two hydrogen bonds (Figure 2a). Hovorun92 assumes that it is possible to have a metastable state having a third hydrogen bond. Rare tautomeric forms of thymine T*4 (Figure 2e) and T*5 (Figure 2f) are possible only in the case when such short-lived semi-open states are formed. It is easy to see that the mechanism of formation of rare tautomeric forms of paired bases of DNA depends only on the properties of hydrogen bonds and properties of DNA molecules. Consequently, it will be true under the action of a DNA molecule of any mutagens. Since the mutagen causes any damage to the DNA molecule, it exhibits excitation energy. This energy is absorbed by one of the DNA bases this lead to the excitation of the electron-vibrational states. At the thermal relaxation of the excitation energy it will cause fluctuations in the lengths of the hydrogen bonds between paired bases. Change in lengths of the hydrogen bonds can lead to changes of tautomeric states of DNA bases. Under certain conditions, the formed rare tautomeric forms of DNA bases will be stable.

Figure 2 Possible tautomeric state of thymine and adenine: a) – canonical thymine – adenine base pair; b) – f) rare tautomeric states of thymine and adenine.73
Polymerase tautomeric models for targeted base substitution mutagenesis during error prone and SOS synthesis of double stranded DNA, containing cis-syn cyclobutane thymine dimers
SOS induction allows DNA synthesis to occur even on templates containing the dimers94 replication on a damaged DNA template, however, results in mutations.93,94 Numerous experimental data61,62 support conclusion that during the error-prone or SOS synthesis or a “sliding clamp” involved in the synthesis95–102 the canonical bases, which can form hydrogen bonds with bases of the template DNA, are incorporated opposite cyclobutane pyrimidine dimers.76 Thus, the following conclusions can be drawn. An exonuclease-free DNA polymerase or a DNA polymerase with the 3¢®5¢-exonuclease activity suppressed by the sliding clamp can incorporate bases opposite DNA bases that are in rare tautomeric forms. Such bases can form hydrogen bonds with the bases of the template DNA, and canonical bases are, as a rule, incorporated. Based on these conclusions, let us now consider the mechanisms for the formation of targeted transitions and transversions during the SOS-replication of double-stranded DNA containing cis-syn cyclobutane thymine dimers. Cyclobutane pyrimidine dimers and (6-4) adducts cause substutution mutations.20,21,103 Experimental data on the operation of a large number of polymerases which incorporate the bases opposite abasic sites, cyclobutane pyrimidine dimers and (6-4) photoproducts have been analyzed, such polymerases as iota (ι), kappa (κ), T7, Dpo4, polξ, DinB family, Rew I, polV, polIV, polα, Tag(pol I family), HIV reverse transcriptase, polδ have been analyzed in Ref28 The following analysis indicates how DNA containing dimers with one or two bases in the rare tautomeric forms shown in Fig. 2 are replicated during error-prone or SOS synthesis. Canonical tautomeric forms of guanine can be incorporated opposite T*1 (Figure 3a). In this case, A:T®G:C transition will result. The insertion of a canonical tautomeric form of thymine opposite T*1 (Figure 3b) produces homologous A:T®T:A transversion. The rare T*1 tautomer cannot form hydrogen bonds with canonical tautomers of cytosine or adenine for steric reasons. The rare T*4 thymine tautomer is capable of forming three hydrogen bonds with cytosine (Figure 3e). This pairing results in A:T®C:G transversion. T*5 can form two hydrogen bonds with cytosine (Figure 3c) and two hydrogen bonds with thymine (Figure 3d). These pairings result in homologous A:T®T:A transversion and in A:T®C:G transversion, respectively.72,77 Nevertheless, polymerases do incorporate mismatched nucleotide base pairs at low frequency.104 Recently, a T•G mismatch has been observed to adopt a canonical base-pair structure in a polymerase, due to an ionization event, demonstrating that noncanonical hydrogen-bonding pattern can arise in a polymerase.61 The results61 suggest a catalytic mechanism for misinsertion and mismatch extension that is in common with correct incorporation, and they support Watson and Crick’s original idea that spontaneous base substitutions, in this case A•T to G•C transition mutations, may result from mismatches shaped like correct base pairs. Wang et al.62 shown that under conditions which stabilize an enzyme conformation that places a nucleotide at the site of incorporation, the C•A mismatch adopts a tautomeric cognate base-pair shape.

Figure 3 Possible base pairs formed between bases in rare and canonical tautomeric conformations. (a) T*1 and G; (b) T*1 and T; (c) T*5 and C; (d) T*5 and T; (e) T*4 and C; (f) T*4 and A.72
Polymerase tautomeric models for targeted insertional mutagenesis during error prone and SOS synthesis of double stranded DNA, containing cis-syn cyclobutane thymine dimers
Insertions are the structural DNA changes wherein one DNA strand becomes longer than the other as a result of an insertion of a number of nucleotides.105 Insertions may be targeted and untargeted types.106 Frameshifts mutations often account for approximately one-third of all mutations.107 Frameshift mutations most commonly arise in DNA sites with a homogenous nucleotide composition, such as monotonous runs of G-C or A-T pairs or sequences with alternating A-T and T-A pairs. Now it is still unclear how frameshift mutations arise at cyclobutane pyrimidine dimers. The Streisinger model108 is now the best-grounded model of frameshift mutations.109–111 suggesting gaps and DNA strand slippage during synthesis as the causes of mutations. Consider the formation of longer insertions. A DNA site is assumed to have a homogenous nucleotide composition and carry two cis-syn cyclobutane pyrimidine dimers in one strand. One dimer is the cis-syn cyclobutane thymine dimer TT2*, and the other is a cyclobutane pyrimidine dimer whose bases occur in the canonical tautomeric forms (Figure 4) (Figure 5a). A structural analysis is performed for the incorporation of DNA nucleotides opposite to T2* (Figure 4) to identify the canonical nucleotides that can be added opposite to T2* to allow hydrogen bonding of the two bases. Canonical thymine cannot be added opposite to T2* by DNA polymerase because of repulsion between the hydrogen H3 of the canonical thymine and H3 of T2* (Figure 4c). Adenine cannot be added because of repulsion between H¢6 of adenine and H¢6А of T2* (Figure 4d). Cytosine incorporation is prevented by repulsion between H4¢ of cytosine and H¢6А of T2* (Figure 4e), and guanine incorporation is prevented by repulsion between H¢1 of guanine and H3 of T2* (Figure 4f). That is, none of the canonical bases can be incorporated opposite to T2*.79 A one-nucleotide gap arises opposite to a cis-syn cyclobutane dimer TT2* (Figure 5b) as a result of translesion synthesis driven by modified E.coli DNA polymerase III or mammalian DNA polymerase δ or ε or specialized (mammalian Polη or Polζ or E.coli DNA polymerase IV or V) DNA polymerases. As was demonstrated experimentally, such a gap arises during DNA synthesis when the template contains an abasic site, leading to a one-nucleotide deletion.112 The nascent DNA strand may slip (Figure 5c) because a bend forms in the site containing cyclobutane pyrimidine dimers and the hydrogen bonds are disrupted.113-116 Since the template in question has a homogenous nucleotide composition, the growing DNA strand may form a small loop (Figure 5d) via pairing with the adjacent nucleotide of the opposite strand. The growing strand is extended by only one nucleotide in this case. The gap increases to two nucleotides; and when it is filled in by constitutive DNA polymerases, a targeted one-nucleotide insertion arises (Figure 5e).79 Specialized or modified DNA polymerases drive DNA synthesis. Hence, a one-nucleotide gap arises opposite to the cis-syn cyclobutane thymine dimer TT2*. The gap arises opposite to T2*. The end of the DNA strand may slip, especially when another (any) cyclobutane dimer occurs in the vicinity of the dimer TT2* because the strand bends and hydrogen bonds are disrupted opposite to such dimers. Since the template region is structurally homogeneous, the end of the growing strand may form hydrogen bonds with a neighbor region to produce a large loop. The resulting large gap is usually filled in by constitutive DNA polymerases, leading to insertion of several nucleotides. A targeted insertion forms in this case. The above mechanisms of insertions agree with the Streisinger model.108

Figure 4 Structural analysis of the potential pairing of the tautomeric form TT2* with the canonical DNA bases. (a) Canonical pair A-T. (b) Pair A2*-T2*, wherein the bases are in the rare tautomeric state. The possibility of T2* pairing is structurally analyzed for the canonical DNA bases (c) thymine, (d) adenine, (e) cytosine, and (f) guanine.79
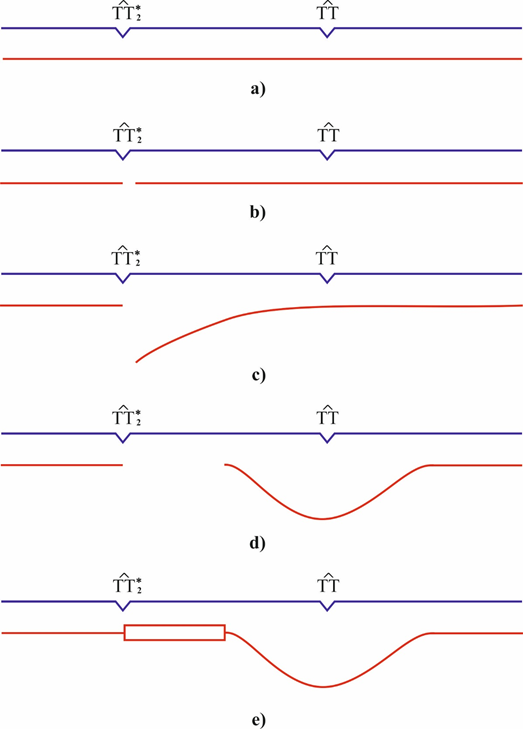
Figure 5 Generation of a targeted insertion of several nucleotides. (a) A DNA site contains the cis-syn cyclobutane dimers TT2* and TT. (b) A one-nucleotide gap arises opposite to the cyclobutane dimer TT2*. (c) The end of the growing DNA strand slips, and (d) a loop forms. (e) The large gap is filled in to produce a targeted insertion of several nucleotides.79
Polymerase tautomeric models for targeted deletional mutagenesis during error prone and SOS synthesis of double stranded DNA, containing cis-syn cyclobutane thymine dimers
The polymerase tautomeric model for targeted deletions (frameshift mutations) caused by cis-syn cyclobutane thymine dimers.80 A one-nucleotide gap arises opposite to a cis-syn cyclobutane dimer TT2* (Figure 6b) as a result of translesion synthesis driven by modified or specialized DNA polymerases. As was demonstrated experimentally, such a gap arises during DNA synthesis when the template contains an abasic site, leading to a one-nucleotide deletion.112 The site in nascent DNA strand may be lost (Figure 6c) because a bend forms in the site containing cyclobutane pyrimidine dimers and the hydrogen bonds between the bases are broken.88, 89, 113–116 A DNA site containing the cis-syn cyclobutane dimers TT2*, may form a loop as shown in (Figure 6e). The resulting smaller gap is usually filled in by constitutive DNA polymerases (Figure 6f), leading to the precipitation of several bases (deletion formation).80 One nucleotide deletions appear most often. In this case, one nucleotide falls. This deletion may cause a one cis-syn cyclobutane dimer TT2*.80 The mechanisms of deletions agree with the Streisinger model.108
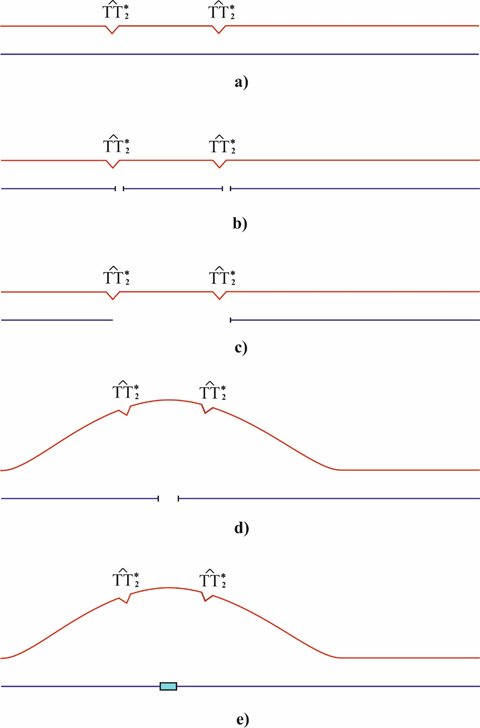
Figure 6 Generation of a targeted deletion of several nucleotides. a) A DNA site contains the cis-syn cyclobutane thymine dimers TT2*; b) a post replicative gap arises opposite to cis-syn cyclobutane dimers TT2*; c) post replicative gap is filled using modified or specialized DNA polymerases. One-nucleotide gaps arise opposite to the cis-syn cyclobutane thymine dimers TT2*; d) site of the DNA strand is lost; e) a loop forms; f) the gap is filled. An insertion of several nucleotides formed, but smaller than the fallen DNA site. A targeted deletion of several nucleotides is formed.80
Polymerase tautomeric models for targeted complex insertions during error prone and SOS synthesis of double stranded DNA, containing cis-syn cyclobutane thymine dimers
Complex mutations are frameshift mutations with an adjacent base substitution.117–119 A polymerase-tautomeric model for targeted complex insertions caused by the cis-syn cyclobutane thymine dimers were suggested.42,81 Cis-syn cyclobutane dimers TT2* may result in targeted insertion.42,79 A one-nucleotide gap forms opposite cis-syn cyclobutane dimers TT2* (Figure 7b). The nascent DNA strand may slip because a bend forms and the hydrogen bonds between the bases are broken88, 89,113–116 (Figure 7c). Since the template site is structurally homogeneous, the end of the growing strand may form hydrogen bonds with a neighbor site to produce a large loop (Figure 7d). The resulting large gap is usually filled in by constitutive DNA polymerases (Figure 7e), leading to targeted insertion of several nucleotides. Four nucleotides will be incorporated. One nucleotide will be incorporated opposite thymine T2*. But a one-nucleotide gap was opposite cis-syn cyclobutane dimers TT2* (Figure 7b). Consequently, an additional three nucleotides incorporated, the insertion of three nucleotides is formed. A DNA site containing the cis-syn cyclobutane thymine dimers TT5*, TT2*, TT1* and TT5* where the thymine molecules in the rare tautomeric forms, corresponding to (Figure 2). They are in one of DNA strands. The opposite DNA strand containing the molecules of adenine A*5, A*2, A*1, and A*5, in the rare tautomeric forms and conforming (Figure 2). A one-nucleotide gap arises opposite the cis-syn cyclobutane thymine dimers TT2* (Figure 7b). Thymine or guanine is inserted opposite T5* in first cis-syn cyclobutane thymine dimers TT5*. Guanine or thymine is inserted opposite T1*. Cytosine or thymine is inserted opposite T5* in second cis-syn cyclobutane thymine dimers TT5* (Figure 7b). The end of the growing DNA strand slips (Figure 7c). d) A loop forms (Figure 7d). The large gap is filled in to produce a targeted insertion of several nucleotides (Figure 7e). The DNA site ATTGTTTTTTTTTATTGT consisting 18 nucleotides has been replaced by the DNA site ATA(G)GTTTTTTTTTTC(A)TAТG(A)GT consisting 21 nucleotides (Figure 7f). Thus, the DNA site ATTGTTTTTTTTTATTGT consisting of 18 nucleotides is replaced by DNA site ATA(G)GTTTTTTTTTTC(A)TATG(A)GT consisting of 21 nucleotides. The same cis-syn cyclobutane thymine dimers can lead to several targeted base substitution mutations.76 For this reason, different targeted complex mutations may appear on the same site of DNA. The DNA site ATTGTTTTTTTTTATTGT consisting of 18 nucleotides may be replaced by 8 DNA site consisting of 21 nucleotides. There are ATAGTTTTTTTTTTCTATGGT, or ATAGTTTTTTTTTTATATGGT, or ATAGTTTTTTTTTTCTATAGT, or ATAGTTTTTTTTTTATATAGT, or ATGGTTTTTTTTTTCTATGGT, or ATGGTTTTTTTTTTATATGGT, or ATGGTTTTTTTTTTCTATAGT, or ATGGTTTTTTTTTTATATAGT DNA sites.42,81

Figure 7 Generation of a targeted complex insertion of several nucleotides.42
Polymerase tautomeric models for radiation induced genomic instability: targeted delayed base substitution mutations during error prone and SOS synthesis of double stranded DNA, containing cis-syn cyclobutane thymine dimers
The conventional view of radiation mutagenesis is that radiation induces most mutations in cells shortly after irradiation.120 Delayed mutations are mutations occur in the progeny of the irradiated cell after many generations of cell division.18 Ultraviolet light induces delayed mutations.23 The delayed mutations are usually point mutations.121 The genome instability results in base substitutions or deletions or insertions of a few nucleotides. Radiation-induced genome instability is a critical early event in the multi-step sequence leading to radiation-induced cancer.122 Radiation-induced genome instability is the process whereby gene mutations increases.18,123,124 Delayed effects include hyper mutation, hyper-homologous recombination, chromosome instability and reduced clonogenic survival (delayed death).125 Delayed mutations and untargeted mutations are two features of genomic instability.126 Although radiation-induced genomic instability has been studied for years, questions regarding the time course of formation and mechanism of induction of delayed mutations remain to be answered.18,127,128 I proposed polymerase-tautomeric models for radiation induced genomic instability: targeted delayed base substitution mutations during error prone and SOS synthesis of double-stranded DNA, containing cis-syn cyclobutane cytosine86 and thymine49,83 dimers. In order to determine which of the canonical bases will be inserted by the SOS-modified DNA-polymerase opposite cis-syn TT3* cyclobutane thymine dimers (Figure 8), consider the constraints on the formation of hydrogen bonds between the bases of the template DNA and the inserted bases. During SOS synthesis of DNA containing dimers, nucleotide bases are inserted opposite the dimers without the removal of the dimer-containing sites. This is only possible when the DNA-polymerase is pressed on the DNA by the “sliding clamp”, obstructing the operation of exonucleases, or when the synthesis involves specialized DNA polymerases, such as E. coli polymerase V or IV, or when the specialized DNA-polymerase is pressed on the DNA by the “sliding clamp”. The rare T3* thymine tautomer is capable of forming one H-bond with canonical adenine (Figure 8c). But T3* can form two H-bonds with canonical guanine (Figure 8d) and one H-bond with canonical cytosine (Figure 8e) and one H-bond with canonical thymine (Figure 8f). Consider a DNA site with a cis-syn TT3* cyclobutane thymine dimers. Let other cis-syn cyclobutane pyrimidine dimers are located quite far from it. Since the damage is only one, the synthesis through the damage will go quite quickly and with high accuracy. For example, the synthesis will be carried out using Pol III DNA polymerase of Escherichia coli or eukaryotic DNA polymerase δ. If a wrong nucleotide is inserted opposite the cis-syn cyclobutane thymine dimer TT3*, the erroneous nucleotides can be removed by 3'®5'-exonucleases. Therefore, with a high probability adenine will be inserted opposite thymine TT3*. In this case the strand of DNA containing cis-syn cyclobutane thymine dimers TT3* does not result in mutations. So many cycles of DNA replication can continue. However, if further DNA synthesis will involve DNA polymerases having a low fidelity of synthesis, there may be base substitution mutations. Moreover, they may be formed through many cycles of replication after DNA has been damaged. Consequently, these are the delayed mutations. Cis-syn cyclobutane thymine dimers TT3* may result in targeted delayed transitions T-A®C-G, targeted delayed transversions T-A®G-C and T-A®A-T.49,83 Opposite canonical thymine cytosine can be incorporated only. Canonical cis-syn cyclobutane thymine dimers TT may result in targeted delayed transversions T-A®G-C only.49
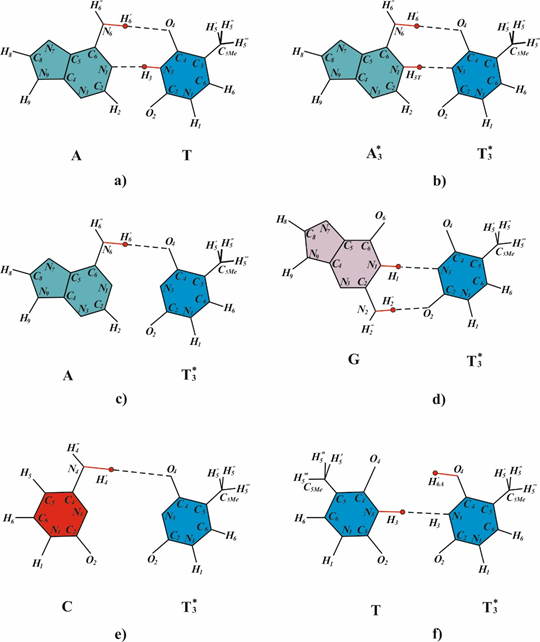
Figure 8 Rare tautomeric form for thymine T3* and possible base pairs formed between thymine in rare T3* tautomeric forms and bases in canonical tautomeric conformations: a) canonical adenine-thymine base pair; b) rare tautomeric forms of thymine T3* and A3* of adenine; c) – f) possible base pairs formed between thymine in rare T3* tautomeric forms and bases in canonical tautomeric conformations: c) T3* and adenine; d) T3* and guanine; e) T3* and cytosine; f) T3* and thymine.49
Polymerase tautomeric models for radiation induced bystander effects: untargeted mutagenesis during error-prone and SOS synthesis of double-stranded DNA, containing cis-syn cyclobutane thymine dimers
I proposed and develops the polymerase-tautomeric model for radiation-induced bystander effects.87 The bystander effects are defined as the induction of cellular damage in unirradiated cells, induced by irradiated cells in the surrounding area.18,24 Over 90% of the mutations arising in bystander cells were point mutations.129,130 Radiation-induced bystander effects include only untargeted mutations.
Polymerase tautomeric models for untargeted base substitution mutations
Let us consider a site of DNA, on which cis-syn cyclobutane thymine dimers with bases in rare tautomeric forms appeared in both strands of DNA close to each other. Besides let us consider a site of DNA, on which in a small neighborhood of cyclobutane pyrimidine dimers with bases in the canonical tautomeric forms pairs base of adenine-thymine in rare tautomeric forms are formed. These sites are synthesized as a result of error-prone or SOS synthesis. Structural analysis indicates that canonical tautomeric forms of thymine cannot be incorporated opposite A1*. But canonical tautomeric forms of cytosine or adenine can be incorporated opposite A1*. Rare A1* tautomer of adenine may result in a untargeted transition A-Т®G-С or a untargeted homologous transversion А-Т®Т-А.87 Molecule of thymine can be inserted opposite A2* and A4*87 molecule of adenine can be inserted opposite T3*49 it is likely they will not result in mutations. The rare A3*, A5* and T2* (Figure 4)79,80 tautomers do not form hydrogen bonds with any canonical tautomers of DNA bases. So they cannot result in the base substitution mutations. Rare T1* tautomer of thymine may result in A-Т®G-С untargeted transition or А-Т®Т-А untargeted homologous transversion (Figure 3).87 Molecules of the thymine in rare tautomeric form T4* may result in transversion A-Т® С-G only.87 Rare T5* tautomer can result in transversion A-Т® С-G or homologous transversions А-Т®Т-А (Figure 3).87
Polymerase tautomeric models for untargeted insertions
The polymerase-tautomeric model for the mechanism of the formation of untargeted insertions caused by thymine and adenine in certain rare tautomeric forms is proposed.85 Structural analysis indicates that opposite the rare tautomers of thymine T2* (Figure 4) and adenine A3* it is impossible to insert any canonical base so that hydrogen bonds between the rare tautomers of thymine T2* or adenine A3* and the canonical bases of DNA are formed. In doing so, author based on the following facts. Specialized or modified DNA polymerases can incorporate bases opposite DNA bases that are in rare tautomeric forms. Such bases can form hydrogen bonds with the bases of the template DNA, and canonical bases are, as a rule, incorporated. The error-prone and SOS synthesis of double-stranded DNA containing the rare tautomers T2* or A3* in one of its strands is considered. This synthesis will result in one-nucleotide gap opposite thymine T2* or adenine A3*. On DNA sites with a homogenous nucleotide composition the end of the nascent DNA strand can slip. The end of the growing strand may form hydrogen bonds with a new site. A loop can form. As a result, the daughter strand is elongated leading to untargeted insertion (Figure 9).85 The polymerase tautomeric model for bystander effects is able to explain the mechanisms formation for untargeted base substitution mutations and untargeted insertions.

Figure 9 Formation of an untargeted insertions from several nucleotides. a) a DNA site containing 2 cyclobutane pyrimidine dimers and, next to them, the rare tautomer of thymine T2* or the rare tautomer of adenine A3*; b) a one-nucleotide gap arises opposite thymine T2* or adenine A3*; c) the end of the growing DNA strand slips; d) a loop is formed; e) the gap is filled in. In result an untargeted insertion of several nucleotides is produced.85
The modern theory of a UV-mutagenesis cannot exhaustively explain many phenomena, including the reason of formation of targeted, delayed and untargeted mutations. The polymerase-tautomeric model for targeted mutagenesis is able to explain the mechanisms formation for targeted substitution mutations,42,76 targeted insertions,42,79 targeted deletions42,80 and targeted complex insertions.42,81 The polymerase-tautomeric models for radiation induced genomic instability is able to explain the mechanisms formation for targeted delayed base substitution mutations.42,86 The polymerase-tautomeric model for radiation-induced bystander effects is able to explain the mechanisms formation for untargeted substitution mutations84 and untargeted insertions.85 Untargeted mutations appear opposite DNA bases in rare tautomeric forms located, for example, near (2-3 bases) from cyclobutane dimers. It is observed a thymine-guanine (T•G)61 and a cytosine-adenine (C•A)62 base pairs with one of the bases in rare tautomeric forms in the active site of DNA polymerases. This results are an experimental verification of polymerase-tautomeric models.
None.
The author declares that there is no conflicts of interest.

©2019 Grebneva. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.