International Journal of
eISSN: 2573-2889


Research Article Volume 6 Issue 1
Department of Ecotourism and Wildlife Management, Federal University of Technology, Nigeria
Correspondence: Ajayi GO, Department of Ecotourism and Wildlife Management, Federal University of Technology, Akure, Nigeria
Received: July 22, 2022 | Published: April 18, 2023
Citation: Ajayi GO, Ogunjemite BG. Genetic characterization of patas monkey (Erythrocebus patas) in new-bussa, niger state, Nigeria. Int J Mol Biol Open Access. 2023;6(1):1-6. DOI: 10.15406/ijmboa.2023.06.00145
This study was carried out to identify, organize and characterize the Patas monkey (Erythrocebus patas) genome. Minimally invasive technique was used to obtain the blood samples for analysis. The blood samples were taken with the assistance of a professional veterinary doctor. They were transferred immediately to blood tubes which contain 95% ethanol for preservation and then stored in refrigerator before laboratory analysis. Genetic analyses revealed that the mitogenome was 16,678 base pair (bp) in length, with an overall base composition of 31.8% A, 25.8% T, 29.7% C, and 12.7% G. The A+T content (57.6%) was greater than the G+C content (42.4%). Phylogenetic analysis was based on the COI sequences of 9 individuals from different locations. The newly sequenced individuals are from Benin Republic, South West, Nigeria and North Central, Nigeria, West Africa; while other sequences were downloaded from the National Center for Biotechnology Information (NCBI database). The nucleotide diversity shows both the frequency and differences between haplotypes, hence, it is a more suitable parameter to estimate the genetic diversity in populations than haplotype diversity. The Kimura-2-parameter (K2P) was used to estimate the genetic distances among the aligned sequences. The study indicates significant genetic differentiation among all of the geographical locations which indicates high genetic distances. The findings revealed a relatively low frequency polymorphisms and absence of variants of alleles among the population of patas. This exemplified a stable demographic history with a stable population size. Hence, the study provides a useful database for analyzing the phylogenetic relationship of Erythrocebus patas. However, further genetic study is recommended for improved conservation of primates in Nigeria.
Keywords: phylogeny, phylogeography, primates, genetic diversity, mitogenome
Africa is a continent of keen interest in terms of global primate conservation. In the study carried out by Ogunjemite1 it was noted that many primate species are threatened and population are on the decline in Nigeria which is as a result of anthropogenic activities. The non-human primates serve as a basic component of tropical biodiversity, contributing to ecosystem health and forest regeneration.2 The Primates are in the Order of animals which comprises of Prosimians, Monkeys, Apes and Humans. Mitani et al3 noted that primates occupy a wide range of habitat types, their ecology varies, and they display a broad array of social and sexual systems. Patas monkey (Erythrocebus patas) lives in African savanna woodlands and grassland habitats, they have a locomotor system that allows them to run fast, most probably to avoid predators. The species has a wide distribution across Sub-Saharan Africa from the Western tip of Senegal to East Africa.4
Genetics is a branch of biology concerned with the study of genes, genetic variation, and heredity in organisms.5 Conservation genetics play a great role in strategic development of short and long term preservation of biodiversity.6 Molecular characterization helps to determine the breeding behavior of species, individual reproductive success and the existence of gene flow, which is, the movement of alleles within and between populations of the same or related species, and its consequences.7 Information on the genetic structure of animal populations can allow us deduce mechanisms shaping their social organization, dispersal, and mating system. Advances in conservation genetics and genomic techniques have led to several findings in the fields of biology and ecology.9 The characterization of genetic diversity pattern at intra- and inter-population levels is a fundamental requirement for the establishment of programs aimed at conservation of genetic resources.10 Molecular characterization provides reliable information for assessing the amount of genetic diversity.11 Phylogenetics and population genetics are well established lines of research that have made significant contributions to the understanding of primate behavior.12
The study by Ojo et al.12 revealed that there are many causes of habitat loss in Borgu Local Government Area due to activities such as logging, overpopulation, urbanization and has led to fragmented population of species. Hence, for conservation priorities, the study of genetic characterization of Red Patas monkey will provide baseline information for this fragmented population of primate species in this savanna ecozone of Nigeria. Hence, the declining population of biodiversity requires the approach of conservation genetics in the assessment of genetic variability and the maximization of genetic diversity. This paper aims to identify, organize and characterize the Patas monkey (Erythrocebus patas) genome in New-Bussa, Niger State, Nigeria.
Study area
The study was carried out in Niger State, Nigeria. Nigeria is situated on the West Coast of Africa, lies on latitudes 9.0820°N of the Equator and latitudes 8.6753°E of the Greenwich Meridian.14 Nigeria shares boundaries with The Republics of Benin and Niger in the West, Cameroon in the East, Niger and Chad in the North and the Gulf of Guinea in the South. Nigeria has an area of 923,768.64km2 (Figure 1).
Data collection
Collection permit was obtained from the Conservator General of National Parks Service, Nigeria and the Conservator of Kainji Lake National Park. The permit is necessary for collection of samples needed for the research. The blood samples of Patas monkey (Erythrocebus patas) were collected from Federal College of Wildlife Management, New Bussa and Kainji Lake National Park Head-Offices in New-Bussa and Wawa, Niger State, Nigeria.
Minimally invasive technique was used in getting the samples for laboratory analysis. Invasive genetics is one of the tools for clarifying society and ecology of captive primates. The technique involves the extraction of DNA from blood or tissue samples such that it spares the life of the animal.15 Tissue, blood and semen are the best source to obtain a DNA profile.16 Also, many behavioral, ecological, physiological, and medicinal studies for conservation purposes require the use of blood samples. The logistics of trapping and sampling wildlife vary greatly depending upon the species of interest. Some species are relatively easy to trap and sample, while others are difficult or dangerous.17 Patas Monkeys are known to be active animals, hence; sedatives were used to calm them, relax their muscle and to reduce their stress level. 0.8ml dose of Ketamine was injected into the animal so as to make it unconscious of its environment. Afterwards, the blood samples were taken from the veins in the arm and directly from the heart with the aid of needle and syringe; this was done with the assistant of a professional veterinary doctor. The samples were transferred immediately to blood tubes which contain 95% ethanol to preserve the samples collected and then stored in the refrigerator before laboratory analysis.
Field study
Global Positioning System (GPS), Syringes, Needles, Blood tubes (with prepared ethanol), Plastic bags, Field notebook and Pen, Paper tape, Marker, Camera, and Rubber gloves.
Molecular study
Micro tubes, Polymerase Chain Reaction (PCR) tubes, centrifuge, Hand gloves, reagents, Gel electrophoresis machine, Microwave, lab coat, thermomixer, UV spectrometer, reagents and pipette.
Storage method includes the use of ethanol and ice/refrigerator.
The blood samples of Patas monkey (Erythrocebus patas) was deposited in the Animal Branch of Germplasm Bank of Wild species, Kunming Institute of Zoology, Chinese Academy of Sciences, China.
DNA extraction
DNA was extracted using DNeasy Blood & Tissue Kit (QiaGen); this was done according to the instruction manual and manufacturer’s protocol. This protocol is designed for purification of total DNA from samples and specimens. The DNA purity and concentration were assessed by spectrophotometry using nano drop and also gel electrophoresis. Library construction and sequencing was done in the Southern China DNA Barcoding Center, with Illumina Miseq platform, reads were assembled using Linux-OS SPAdes genome assembler v3.12.0 Bankevich et al.18 with k-mer.19,20
Data analysis
The tRNAs sequences were confirmed using online Search Service tRNAscan-SE Schattner et al.21 and annotated with the online program DOGMA.22 The final genome map was generated using Organellar Genome DRAW.23
DOGMA helps in the verification, to ensure that all protein-coding genes are correctly transcribed, and rRNAs and tRNAs are able to form their typical secondary structure. Other Sequences were downloaded from NCBI GenBank for the construction of its phylogeny. The neighbor joining tree (NJ) and maximum likelihood (ML), were performed in MEGA 7.0 while maximum parsimony (MP) was estimated using PAUP_ (V4.0) Swofford20 for phylogenetic reconstruction comparism.
Molecular Evolutionary Genetics Analysis (MEGA 7.0) was used to compare and align sequences for differences. MEGA is an integrated tool for conducting automatic and manual sequence alignment, inferring phylogenetic trees, mining web-based databases, estimating rates of molecular evolution, and testing evolutionary hypotheses.23
The phylogenetic analysis was conducted to assess the taxonomic relationship of the assembled newly COI sequences of E. patas with other Erythrocebus patas downloaded from the National Center for Biotechnology Information (NCBI), with Chlorocebus sabeaus serving as outgroups. The Phylogenetic tree was constructed with MEGA 7.0 using the Neigbour Joining Tree (NJ). The mtDNA COI sequences were analyzed through Kimura 2 parameter (K2P) by using MEGA 7.0 to infer the genetic distance and monophyletic clustering of the studied taxa.25
The genome organization
The mitogenome was 16,678 bp in length, with an overall base composition of 31.8% A, 25.8% T, 29.7% C, and 12.7% G. The mitogenome consists of 13 protein-coding (PCGs), 2 ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and 1 control region (D-loop) (Table 1). Most of the PCGs initiation codons were ATG except for ND3 and ND2 with a slight difference in the initiation codon of ATT and ATA respectively. In addition, 9 of the 13 protein-coding genes had completed termination codon CCT and TAA, while ATP6, ND4, ND3 and CYTB genes terminate with incomplete stop codon (T-). A total of thirty-seven (37) gaps/overlaps were identified among the genes. The Heavy strand (H strand) carries most of the genes, while the remaining gene are encoded on the light strand (L strand).
|
Name |
Type |
Minimum |
Maximum |
Size(bp) |
Direction |
Start codon |
Stop codon |
Gaps+/overlaps-(nt) |
|
tRNA-Phe |
tRNA |
1 |
71 |
71 |
Forward |
0 |
||
|
12S Rrna |
rRNA |
72 |
1017 |
946 |
Forward |
4 |
||
|
tRNA-Val |
tRNA |
1022 |
1091 |
70 |
Forward |
0 |
||
|
16S Rrna |
rRNA |
1092 |
2639 |
1548 |
Forward |
6 |
||
|
tRNA-Leu |
tRNA |
2646 |
2720 |
75 |
Forward |
2 |
||
|
ND1 CDS |
CDS |
2723 |
3677 |
955 |
Forward |
ATG |
CCT |
0 |
|
tRNA-Ile |
tRNA |
3678 |
3745 |
68 |
Forward |
-3 |
||
|
tRNA-Gln |
tRNA |
3743 |
3814 |
72 |
Reverse |
1 |
||
|
tRNA-Met |
tRNA |
3816 |
3883 |
68 |
Forward |
0 |
||
|
ND2 CDS |
CDS |
3884 |
4925 |
1042 |
Forward |
ATT |
CCT |
0 |
|
tRNA-Trp |
tRNA |
4926 |
4991 |
66 |
Forward |
7 |
||
|
tRNA-Ala |
tRNA |
4999 |
5067 |
69 |
Reverse |
1 |
||
|
tRNA-Asn |
tRNA |
5069 |
5141 |
73 |
Reverse |
32 |
||
|
tRNA-Cys |
tRNA |
5174 |
5241 |
68 |
Reverse |
-1 |
||
|
tRNA-Tyr |
tRNA |
5241 |
5306 |
66 |
Reverse |
16 |
||
|
COX1 CDS |
CDS |
5323 |
6891 |
1569 |
Forward |
ATG |
TAA |
-28 |
|
tRNA-Ser |
tRNA |
6864 |
6932 |
69 |
Reverse |
3 |
||
|
tRNA-Asp |
tRNA |
6936 |
7003 |
68 |
Forward |
1 |
||
|
COX2 CDS |
CDS |
7005 |
7688 |
684 |
Forward |
ATG |
TAA |
44 |
|
tRNA-Lys |
tRNA |
7733 |
7798 |
66 |
Forward |
1 |
||
|
ATP8 CDS |
CDS |
7800 |
8000 |
201 |
Forward |
ATG |
TAA |
-40 |
|
ATP6 CDS |
CDS |
7961 |
8641 |
681 |
Forward |
ATG |
T(AA) |
-1 |
|
COX3 CDS |
CDS |
8641 |
9424 |
784 |
Forward |
ATG |
CCT |
0 |
|
tRNA-Gly |
tRNA |
9425 |
9492 |
68 |
Forward |
0 |
||
|
ND3 CDS |
CDS |
9493 |
9838 |
346 |
Forward |
ATA |
A(AT) |
0 |
|
tRNA-Arg |
tRNA |
9839 |
9903 |
65 |
Forward |
0 |
||
|
ND4L CDS |
CDS |
9904 |
10200 |
297 |
Forward |
ATG |
TAA |
-7 |
|
ND4 CDS |
CDS |
10194 |
11571 |
1378 |
Forward |
ATG |
C(TT) |
0 |
|
tRNA-His |
tRNA |
11572 |
11639 |
68 |
Forward |
0 |
||
|
tRNA-Ser |
tRNA |
11640 |
11698 |
59 |
Forward |
0 |
||
|
tRNA-Leu |
tRNA |
11699 |
11769 |
71 |
Forward |
6 |
||
|
ND5 CDS |
CDS |
11776 |
13581 |
1806 |
Forward |
ATG |
TAA |
0 |
|
ND6 CDS |
CDS |
13582 |
14100 |
519 |
Reverse |
CAT |
CCT |
0 |
|
tRNA-Glu |
tRNA |
14101 |
14169 |
69 |
Reverse |
4 |
||
|
CYTB CDS |
CDS |
14174 |
15314 |
1141 |
Forward |
ATG |
T(CT) |
0 |
|
tRNA-Thr |
tRNA |
15315 |
15381 |
67 |
Forward |
2 |
||
|
tRNA-Pro |
tRNA |
15384 |
15451 |
68 |
Reverse |
0 |
||
|
D-loop |
15452 |
16676 |
1225 |
Forward |
Table 1 Genome Organization
Phylogenetic analysis
Phylogenetic tree is based on the COI sequences of 9 individuals from different locations. The newly sequenced individual E1 and E4 are from Benin Republic and South West Nigeria and E2 and E3 from North Central, Nigeria, West Africa (Table 2). Others were downloaded from the National Center for Biotechnology Information (NCBI) (Table 2). To validate the reliability of the sequence, comparison was made with other mitochondrial sequences from Erythrocebus species. The phylogenetic position was estimated from complete mtDNA sequences. The phylogenetic placement (Figure 2) shows that the individuals consist of two major groups with six clades (A-F). Clade consists of a monophyletic group of species that shows single common ancestors. Each pair consists of a clade which reveals their closeness or relatedness, that is, they share a recent common ancestor. However, they all share a common ancient ancestors which serve as an outgroup (clade F).
|
S/N |
Sequence name |
City/Country |
Region |
Longitude/Latitude |
Type of sample |
|
1 |
JF444300.1_Erythrocebus_patas_voucher_
|
Ontario, Canada |
East |
79° 11' 9.204'' E 43° 49' 3.7164'' N |
Tissue, Skin, Skeleton, Wet specimens |
|
2 |
LC144609.1_Erythrocebus_patas_mitochondrial
|
Nagoya, Japan |
Central Japan |
136.981271° E 35.157447° N |
Tissue |
|
3 |
KJ192794.1_Erythrocebus_patas_isolate_EpatT1273 _cytochrome_oxidase_subunit_I_(COI)_gene_partial_ cds_mitochondrial |
Ibadan, Nigeria |
South West |
4° 07 ʹ E 7° 20ʹ N |
Processed, stall of smoked meat, smoked, beheaded species
|
|
4 |
KJ192793.1_Erythrocebus_patas_isolate_EpatT1266_ |
Ibadan, Nigeria |
South West |
4° 07 ʹ E 7° 20ʹ N |
Processed, stall of smoked meat, smoked, beheaded species |
Table 2 Sequence DNA of Erythrocebus patas
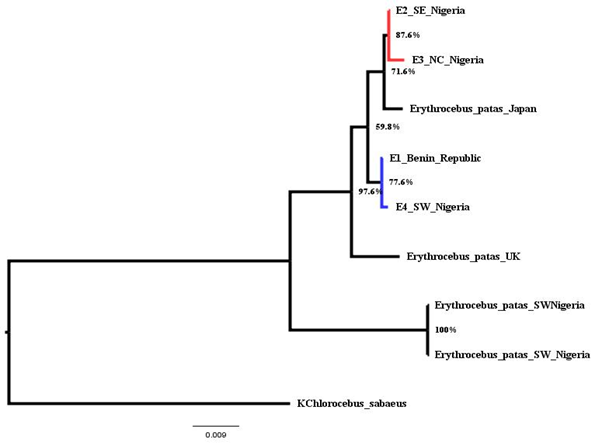
Figure 2 Phylogenetic Relationships among Patas Monkey and Outgroup Taxa based on Complete Mitochondrial Genome Sequences.
Evolutionary sequence divergence
Moderate to high intraspecific genetic distances were observed among lineages. The mean p-distance between different geographical group ranged from 0.014 to 0.028 (Table 3). Highest intraspecific K2P distance of 0.031 (within group) was observed in Nigeria geographical location, while Benin Republic had the lowest intraspecific genetic distances (0.014: Table 3). Our analyses showed significant genetic differentiation among all of the geographical location. However, the sample size might be an influence on the variation within and among groups.
|
Between groups |
Within groups |
||
|
Nigeria |
0.031 |
||
|
Japan and UK |
0.028 |
0.016 |
|
|
Benin Republic |
0.022 |
0.014 |
n/c |
Table 3 Evolutionary sequence divergence of pairwise (p-distance) between and within clades of e. patas from different geographical location
n/c: not able to calculate due to single sequence
Population genetic polymorphism
Table 4 above shows the genetic diversity of individual of E. patas from Nigeria with all sequences. Southwestern Nigeria has the highest number of individual and haplotypes (h=3) while Southcentral and Northcentral has the lowest and same number (n=1). The result further clarified that the haplotype diversity of Southwestern Nigeria ranges from 0.002 to 0.12, Southcentral 0.001 to 0.10 and Northcentral 0.001 to 0.11. The mean haplotype diversity overall populations was 0.004.
|
Sample population |
N |
H |
hd ±SD |
pi ±SD |
|
Genetic diversity ofE. patas Individual from Nigeria |
||||
|
Southwestern |
3 |
3 |
0.002± 0.12 |
0.017 ± 0.04 |
|
Southcentral |
1 |
1 |
0.001± 0.10 |
0.016 ± 0.003 |
|
Northcentral |
1 |
1 |
0.001 ± 0.11 |
0.013 ± 0.00 |
|
Overall genetic diversity |
5 |
5 |
0.004 ± 0.33 |
0.046 ± 0.043 |
Table 4 Population genetic polymorphism based on coi sequence data
N, number of individuals, H, number of haplotypes, hd, gene haplotype diversity (± Standard Deviation), pi, nucleotide diversity (± Standard Deviation)
The nucleotide diversity (pi) shows both the frequency of haplotype and nucleotide differences between haplotypes, hence, it is a more suitable parameter to estimate the genetic diversity in populations than haplotype diversity. The mean nucleotide diversity in the population of E. patas was estimated to be 0.046.
Demographic history of patas
The results Table 5 showed non-significant values (P>0.05) of Fu’s Fs and Tajima’s D for Southwest, Southcentral and Northcentral respectively, which suggests that the population has experienced a stable demographic history. Southwest (3.424; 0.40), Southcentral (2.985; 0.38), Northcentral (1.470; 0.35) population showed non-significant positive values for Fu’s Fs and non-significant negative values (P>0.05) Tajima’s D which is a characteristics of a stable demographic history with a stable population size.
|
Fu’s Fs |
Tajima’s D (p-value) |
|
|
Southwestern |
3.424 (0.40) |
-0.110 (0.51) |
|
Southcentral |
2.985 (0.38) |
-0.065 (0.58) |
|
Northcentral |
1.470 (0.35) |
-0.049 (0.53) |
Table 5 Statistics of neutrality test and demographic history of E. patas
Identification and organization of genes of patas
The mitogenome used for the species identification was 16, 678 base pairs in length with an overall base composition of 31.8% A, 25.8% T, 29.7% C, and 12.7% G. This agreed with previous studies on complete mitochondrial genome sequence for primates carried out by Ayoola and Zhang et al.26 the aim is to identify unidentified species and also to detect unpredicted diversity. Early Studies by authors such as Hebert et al.27 and Bickford et al.28 Reveals the successful identification accuracy of DNA barcodes. Patas mitochondrial genome sequences were successfully amplified and sequenced, this is in line with Zinner et al. (2013) who successfully sequenced ten Papio individuals with length of 16, 858bp.The genome’s A+T content (57.6%) was higher than the G+C content (42.4%); this agreed with the study of Piovesan et al.29 The mitogenome consists of 13 protein-coding (PCGs), 2 ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and 1 control region (D-loop). It has the same gene arrangement and similar codon usage with other primates’ genome.26 The aim of the mitochondrial genome is to infer the evolutionary relationships of patas monkey. The sequencing further helped in the organization of protein-coding information in the context of genomes as a whole.30 A total of 37 gaps/overlaps were identified among the genes. This study clearly affirmed the effectiveness of COI barcodes for the identification of species. Every individual has a distinct COI sequence and COI analysis separated the different species into distinct branches, with species of single common ancestors sharing the same clade. Our findings represent the validity of DNA barcodes in identification of primate species especially closely related patas species and a paradigm shift to focus on conservation of primate through tools like DNA barcodes.
Characterization of patas monkey genome
The Kimura-2-parameter (K2P) was used to estimate the genetic distances among the aligned sequences Candek and Kuntner.16 the COI sequences showed intraspecies and interspecies distance among patas from four countries (Nigeria, Japan, UK and Benin Republic). Moderate to high intraspecific genetic distances were observed among lineages. Highest intraspecific K2P distance of 0.031 (within group) was observed in Nigeria geographical location, this does not agree with studies by (Hebert et al Candek and Kuntner).27 In the study of Hebert et al.27 the COI sequence variation between bird species was on average twenty times larger than that within species while Candek and Kuntner reported that all intraspecific groups of spider species had significantly lower K2P distances compared with interspecific groups.
This study revealed that Benin Republic had the lowest intraspecific genetic distances. There is significant genetic differentiation among all of the geographical location which indicates high genetic distances, this reveals that they are not closely related and do not have a recent common ancestor. Early studies supported this significant difference Hebert et al.27 Nucleotide diversity between the populations of E.patas from Nigeria shows that population from the South West region has the highest genetic polymorphism. The function of DNA barcodes which goes beyond identification to further genetic and evolutionary studies is consistent with the study of Gamaniel and Gwaza9 and Lohman et al.28
The haplotype diversity of population from Southwestern Nigeria ranges from 0.002 to 0.12, Southcentral 0.001 to 0.10 and Northcentral 0.001 to 0.11. The nucleotide diversity (pi) describes both the frequency of haplotype and nucleotide differences between haplotypes, hence, it is a more suitable parameter to estimate the genetic diversity in populations than haplotype diversity. The highest haplotype value was seen in Southwestern Nigeria species which indicates high level of introgression within Southwestern Nigeria’s E. pataspopulation. Northcentral species has the lowest value of nucleotide diversity which can be explained due to low gene flow as explained by Zinner et al.32 that gene flow between populations occurs at differing degrees and at different times.
Evolutionary history of patas
The neighbor joining method was used to construct a phylogenetic tree and individuals are differentiated by their distinct clades within the tree. The clades are grouped according to how similar/related the individuals are; these clades are nested as each branch splits into smaller branches. Each branch represents population through time (known as genetic distance) and the split reveals the evolutionary history. Phylogenetic tree is used for visualization of ancestor-descendant relationships; the more closely, the more related. The constructed tree is able to illustrate the evolutionary history of the various individuals. The tree shows genetic distance (time), which is the measure of genetic divergence between species of population; this is in tandem with the study by Muhammad and Shoaib33 which reveals that molecular approach evaluates the nucleic divergence among species, genus and family.
Evolutionary pattern of complete mitochondrial COI gene sequence of E. patas Benin Republic, E. patas South West Nigeria, E. patas South East Nigeria and E. patas North Central Nigeria was checked with patas Japan, E.patas UK, E.patas South West Nigeria and KChlorocebus sabaeus that were collected from NCBI Nucleotide database. Phylogenetic analysis shows that the E. patas South East Nigeria and E. patas North Central Nigeria species clustered (clade A), having a total of 87.6% bootstrap confidence value are more evolutionarily close to Japan species (clade B); both the sequences were branched as nodein the tree as shown in Figure 2 showing that they share the recent or close-related ancestors. The E. patas Benin Republic, E. patas South West Nigeria species also clustered (clade C), and are more evolutionarily close to the UK species (clade D), which serve as the sister lineage. Both species of patas SW Nigeria clustered and showed a bootstrap confidence value of 100% (clade E): this indicates that they are of the same species, however genetically close to the UK species. The species, KChlorocebus sabaeus (clade F) branched as outgroup and is the most distantly related to all other species (in-groups), although close to both species of E. patas SW Nigeria. However, the outgroup species (KChlorocebus sabaeus) is known to share the same sub-family of Old World Monkeys (Cercopithecinae) with E. patas species.
In the construction of the phylogenetic tree, sequences separated by shorter evolutionary distance will be similar to one another and organisms that have the fewest traits branch off first while organisms with most traits branch off later. The similarity in Clade C can be assumed to be as a result of Seme Border which is a settlement in Lagos Nigeria on the border with Benin; this is consistent with the study of Bonadio34 who stated that patas have larger home range and longer daily range, hence, rapid migration. Clade B and Clade D also showed the genetic presence of patas monkey in Japan and United Kingdom respectively, this does not agree with the study of Honolulu Zoo35 that patas monkeys (patas) live only in Africa. Also, in our mitochondrial phylogeny, we found that geographic close lineages cluster together.
Tajima’s D statistic was used to assess the fit of the data to the standard neutral model of a randomly mating population of constant size. The non-significant values (P>0.05) of Fu’s Fs and Tajima’s D for Southwest, Southcentral and Northcentral respectively, opines that the population has experienced a stable demographic history. The non-significant positive values shown by all regions for Fu’s Fsindicates the deficiency of variants of alleles as would be expected from a recent population bottleneck Ashfaq et al.36 and non-significant negative values (P>0.05) Tajima’s D reflects a relative excess of low frequency polymorphisms, as is seen after a population expansion or genetic hitchhiking probably from a recent bottle neck or founder effect Fischer et al.37 and Ashfaq et al.36 this is also a characteristic of a stable demographic history with a stable population size.
Characterizing patas genome is important because they are threatened with anthropogenic activities, and knowledge about genetic composition among population will guide conservation efforts. This research was able to identify unidentified species and to detect unpredicted diversity; it also gave a useful database for analyzing the phylogenetic relationship of E. patas population in Nigeria and other E.patas species. The genome A+T content and G+C content was compared. Diversity levels based on DNA sequences shows that different population of patas have different demographic histories. Thus, the demographics of patas populations will help with its genetic understanding.
The study indicates significant genetic differentiation among all of the geographical locations which indicates high genetic distances; the differences might have been as a result of evolutionary forces to enhance adaptation and productivity. The findings also reflect a relative excess of low frequency polymorphisms and indicates the deficiency of variants of alleles among the population of patas. This is a characteristic of a stable demographic history with a stable population size.
Our sincere gratitude goes to the Conservator General of the Nigerian National Parks Service, the Conservator of Park, Staff of Kainji Lake National Park and Staff of Federal College of Wildlife Management, New-Bussa, Niger State, Nigeria. Thank you for your warm reception and assistance during the data collection period. We also wish to express our gratitude to Miss Ayoola, Adeola Oluwakemi for her undiluted support during the course of analyses of this study.
The authors declare that there are no conflicts of interest.

©2023 Ajayi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.