International Journal of
eISSN: 2573-2889


Review Article Volume 3 Issue 3
Department of Food science and Technology, Sabaragamuwa University of Sri Lanka, Sri Lanka
Correspondence: Rathnayaka RMUSK, Department of Food science and Technology, Faculty of Applied Sciences, Sabaragamuwa University of Sri Lanka, P.O. Box 02, Belihuloya, Sri Lanka
Received: February 12, 2018 | Published: June 7, 2018
Citation: Dias PGI, Rathnayaka RMSUK. Fluorescence in situ hybridization (FISH) in food pathogen detection. Int J Mol Biol Open Access. 2018;3(3):143-149. DOI: 10.15406/ijmboa.2018.03.00066
Diseases due to food borne microbial pathogens are major public health concern in the world. Different methods are used to detect food borne pathogens from different types of foods to prevent from these diseases. Out of those methods, molecular detection methods have received more concern in the recent past. Fluorescence in situ Hybridization is one of such methods used in the detection of food borne microbial pathogens. The present situation of using Fluorescence in situ Hybridization to detect food borne microbial pathogens is review in this article.
Keywords: fluorescence in situ hybridization, food borne pathogens, molecular detection methods
Food borne diseases (FBDs) are of foremost public health concern in the world. Those are accountable for millions of deaths worldwide and placing a tremendous burden on the socioeconomic status of developing countries. Currently, reliable statistical estimates for the global influence of FBDs are not available. However, diarrhoeal diseases alone, which form a generous percentage of FBDs, kill 1.9 million children globally every year.1 The majority of FBDs are generated by the consumption of foods contaminated with pathogenic bacteria, viruses or parasites. Listeria monocytogenes, Escherichia coli O157:H7, Staphylococcus aureus, Salmonella enterica, Bacillus cereus, Vibrio spp., Campylobacter jejuni, Clostridium perfringens, Shiga toxin-producing Escherichia coli and norovirus are some common examples of such microorganisms.2 Mass scale food processors are generally implementing food quality management systems (e.g. ISO 9001) and food safety management systems (e.g. ISO 22000) throughout their entire food chain which restricting the access of food borne pathogens (FBPs) into particular food products. However, increasing demand for street foods and minimally processed foods yet has the risk. Therefore, detection, identification, characterization, and quantification of FBPs in foodstuffs with greater accuracy, sensitivity and rapidity are an utmost important proactive approach to eliminate FBDs.
Simple, easily adaptable and generally inexpensive culturing methods are traditionally exploited over several decays to detect FBPs. However, those methods are time-consuming (minimum 18 to 24 hours), labor-intensive, sometimes giving falsely negative results, and most importantly less sensitive for fewer microbial counts. Further, viable but non-culturable microbes cannot detect by those methods.3 On occasion, single FBP is enough to cause lethal infections. Hence, culture-independent rapid detection methods are important, particularly in the food industry, as they are able to detect FBP with great accuracy and sensitivity. However, expensiveness, complexity and requirement of expertise knowledge limited the fame of these culture independent methods. Rapid FBP detection methods can be categorized into three groups as Nucleic acid-based (NAB), biosensor-based (BB), and immunolgical-based (IB) methods.2 Among them, NAB methods are widely utilized in FBP detection. DNA and RNA are the two nucleic acids present in the living organisms. DNA sequence is carrying genetic instructions to produce a specific amino acid sequence in a protein. RNA is mediate to synthesize protein according to the genetic instruction of DNA. There are specific and unique nucleotide sequences within FBP cells which are responsible for generating toxic proteins (e.g. shiga toxin by E coli). In NAB methods these nucleotides are identified and read, hence confirm the presence of a particular pathogen. Polymerase chain reactions (PCR), nucleic acid sequence-based amplification (NASBA), loop-mediated isothermal amplification (LAMP) and microarray technology are some examples of NAB methods. Fluorescence in situ hybridization (FISH) also can categorize as a NAB method. However, it has not yet been routinely used to analyze and monitor pathogenic microorganisms in food products. FISH has additional benefits as it visualizes whole cells and targets ribosomal RNAs (or other abundant structures like multi-copy genes), which offers FISH with the ability to distinguish between viable organisms and dead materials. Correspondingly, the result can be obtained within few hours and less effort with high specificity and sensitivity.4 The purpose of this review is to give an overview of the current state of the FISH-testing in FBPs detection on diverse kinds of food matrix.
History
The concept of applying molecular hybridization directly into the cytogenetic material is firstly founded by Pardue & Gall5 and independently John et al.6 The method is called In Situ Hybridization (ISH). In ISH, radioactively labeled single-stranded DNA and 28S RNA are used as probes. In ISH, hybridized probes were observed in silver color through radiograph. The technique allowed detecting nucleic acid sequence inside a cell without disturbing its morphology or integrity. ISH was majorly utilized for detecting the chromosomal disorders and took the clues of several diseases as tumors of a wide range of species. Giovannoni et al.,7 was the first researcher who used ISH for bacteriological detections. In the early 1990s introduction of fluorescence labeling displaced the radioactive labeling. In Delong et al.,8 first used fluorescently labeled oligonucleotides for the uncovering of single microbial cells.9 With compare to radioactive probes, fluorescent probes are safer, give better resolution and do not need additional detection steps. Moreover, fluorescent probes can be labeled with dyes of different emission wavelength, therefore, allowing detection of several target sequences within a single hybridization step.9
Basic elements of FISH
Fluorescently labeled, synthesized nucleotide probe/prime and its complementary DNA/RNA sequence within the target cell are the basic FISH elements. Nucleotide probe can be DNA or RNA or else DNA/RNA mimics (PNA- peptide nucleic acid/ LNA- locked nucleic acid). They are commercially available or else can be prepared in laboratory levels and store under -200C for several months. Specificity, sensitivity, and ease of penetration are the major aspects when selecting a probe. Short probes make easy entree to the target but can carry fewer labels. Therefore, the usual probe contains 15 to 30 base pairs.9 Probes are labeled either directly, by chemically or enzymatically bound to fluorescent nucleotides, or indirectly, by integration of reporter molecules.10 16S or 23S rRNA is the majorly used target nucleotide since it is present in all living cells in fairly high amount.11 They are polygenetic markers and lot of sequence data available for probe designing. However, the less sensitivity of FISH protocols is compelled to utilize nucleic acids present in lower copy numbers, such as mRNA, plasmids or even single copy genes. Diversification of target nucleotides are recognized by genome sequence identification projects.12
Basic steps of FISH
The procedure contains following basic steps (Figure 1). (i) Fixation and permibilzation of the specimens, (ii) hybridization of the probe with a target nucleotide, (iii) wash to remove unbound probes and (iv) visualization and enumeration of the result. However, in food microbiology, further steps for the sample preparation as homogenization, pre-enrichment procedures or bacterial separation might be necessary.4
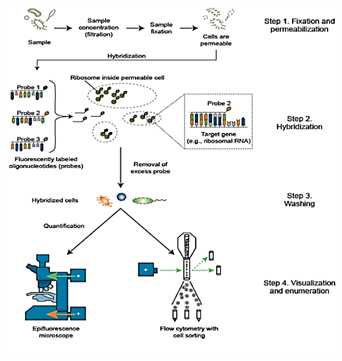
Figure 1 Steps in fluorescence in-situ hybridization
Source: Environmental Molecular Diagnostic, 2017.
Sample preparation
FBPs identification is a predominant challenge in food analysis. Uneven distribution of target microorganism in a little number, the complexity of food matrix, and an enormous number of other competitive micro-flora in the same food create barriers to accurate detection.4 In FISH, there is another restriction as the food components with natural fluorescence activity, like chlorophyll in plant material and hemoglobin in meat are responsible for false results.4,13 Several pretreatments as pre-enrichment and centrifugation are assigned in sample preparation step to expel those obstacles. However, they are not universal or matrix specific.
Fixation and permeabilization of the specimens
Separated cells of interest are fixed into a cleaned glass slide, to preserve their morphology and integrity. Gelatin, Polylysine or Glutaraldehyde coated slides are used to minimize the tissue losses and maximum recovery of oligonucleotides. Precipitation agents such as ethanol or methanol and cross-linking agents such as formaldehyde are some successful fixative agents. Fixation method largely depends on target microorganism. For instance, formaldehyde or paraformaldehyde is recommended for gram-negative bacteria while ethanol, formalin or heat treatment is recommended for gram-positive bacteria.9 Fixation free FISH also grab the attention of researchers.14 It is reported that unfixed FISH minimize the sequencing biases introduced by crosslink between fixative agents and nucleotides as well as avoid possible cell lysis difficulties.14,15 Further, unfixed mammalian tissue has been shown to produce comparatively better imaging results.16 However, unfixed FISH is recommended for fresh samples since it is not preserving the sample as fixation.14 Unfixed oligonucleotide probes of Escherichia coli, Salmonella enterica and Pseudomonas putida were investigated in
several studies, however, yet not in commercial level.14,17,18 Target oligonucleotides are surrounded by proteins, lipids, etc., and that masked the access of fluorescent probes towards the target. Therefore, permeabilization procedures are often required. There is no matrix specific or universal protocol for permeabilization. However, HCL treatment for partial hydrolysis of proteins, detergent treatment for denaturing the lipid membrane components and RNase treatment for inactivate the RNAs and increase the DNA-DNA noise to signal ratio are some common approaches. Further, treatment with unspecific proteases (e.g. Savinase or Proteinase K) is applied to reduce the auto-fluorescence of the food matrices.13
Hybridization, washing and visualization
In hybridization, labeled probes bind with target nucleotide sequence and form ‘probe-target hybrids’. Both should be denatured before hybridization and often formamide is added to reduce the melting point.19 Hybridization temperature, buffer stringency and number and length of nucleotides and number of C-G bonds (stronger than A-T/U bonds) are the factors influence in the process.13 Post-washing procedures remove unbound single stranded DNA as well as nonspecifically bound DNA. Visualization can be performed either by florescence microscope or else flow cytrometry.
Applications of FISH in food pathogen detection
Applications of FISH are largely directed towards medicine and diagnostic field. Rapid and convenient identification of pathogens in the blood or the faeces and cytogenetic examinations to detect chromosomal disorders or tumor cells are the trending topics in molecular based research arena. FISH also utilize in ecology and environmental biology to study the composition, growth and changes of complex microbial communities and biofilms.4,20–22 In addition the method acts as an emerging tool in FBPs detection (Table 1). Nevertheless, this area of FISH possesses large gaps and requirement to upgrade.4 stated that FISH is in its infancy level with compare to molecular techniques as PCR. Some findings within the past decade on FISH in food pathogen detection are summarized in (Table 1). According to the literature, Salmonella spp. and Listeria spp. are the largely investigated pathogens through FISH. Most of the investigations are continued with the aid of artificially introduced (spiked) pathogens. Thus, it is debatable whether the findings can similarly apply to the naturally contaminated food matrix. Liquid, solid, processed and unprocessed matrices were evaluated by FISH, though, the protocols are not matrix specific.
|
Reference |
Pathogen/type of contamination |
Food matrix |
Target probe type |
Remarks (enrichment period/fish methodology/sensitivity) |
|
Salmonella spp. (spiked) |
Lettuce, spinach, parsley, carrot, celery, tomato and sweet corn roots |
Not stated |
confocal laser scanning microscopy (CLSM) |
|
|
Yersinia spp. (spiked) |
minced pork meat |
16S and 23S rRNA LNA |
Use math-FISH in probe development |
|
|
Escherichia coli O157 |
Ground beef Milk (spiked) |
PNA |
100% specificity, 97.22% sensitivity, and 98.33% accuracy |
|
|
Enterobacteriaceae Pseudomonas spp. |
Milk |
16S rRNA |
7 h, multicolor fluorescence in situ hybridization (M-FISH), micro-colony growth method (MMC-FISH) |
|
|
Salmonella spc. |
Powdered infant Formula |
23S rRNA, PNA |
Detection of 1 CFU/10 g in 12 h, 100% sensitivity and specificity |
|
|
Helicobacter pylori |
Bovine milk |
16S rRNA |
PCR amplification, > 4 log CFU/g sensitivity |
|
|
Salmonella spc. (spiked) |
Pork sausages |
4 h enrichment, Pre-enriched samples have high sensitivity |
||
|
30 |
Listeria spp. |
23S rRNA |
14 h required with pre-enrichment step, FISH on filter method to separate microbes |
|
|
Salmonella spp. |
Tomato |
Not stated |
Tape-FISH |
|
|
Salmonella spp. |
Pork sample |
23S rRNA |
Pre-enrichment step enhance the sensitivity of FISH detection |
|
|
Clostridium perfringens |
Food samples |
16S rRNA |
FISH on filter method was employed 9 h test with 2 log CFU/g sensitivity |
|
|
Listeria monocytogenes |
Smoked salmon Mozzarella cheese Julienne cabbage (spiked ) |
16S rRNA |
16 h (cultivation 12 h, fixation 1 h, air-drying 45 min, hybridization 1 h, washing 30 min, counting 30 min) required with aid of FISH on filter method |
|
|
Bacillus cereus |
Milk (spicked) |
16S rRNA |
Flow cytometry, 3 log CFU/mL |
|
|
Salmonella spp. |
Pork meat |
23S rRNA |
16 h pre-enrichment step |
|
|
Mycobacterium avium subsp. |
Portable water |
16S rRNA PNA |
24 h enrichment |
|
|
Salmonella spp. Listaria spp. |
Barley plants (spiked) |
16S rRNA 23S rRNA |
CLSM |
|
|
Campylobacter spp. |
Spiked drinking water |
PNA 16s rRNA |
Membrane filtration technique brightness of the hybridized cells is higher when they are viable |
|
|
3 |
Salmonella spp. |
18 different foods |
23S rRNA |
Convectional culturing methods are more sensitive than FISH, food matrix not interfering the results |
|
Escherichia coli. |
Ikura (Japanies sea food) Minced chicken meat |
16S rRNA, DNA |
FISH on filter method 6 h for enrichment and 1 h FISH assay 2 log CFU/g sensitivity |
|
|
Pseudomonas spp. |
Milk |
16S rRNA |
2 h for FISH assay Flow cytometer |
|
|
Camphylobactor spp. |
Chicken products Spiked and natural |
16S rRNA |
22 h pre-enrichment period |
Table 1 Application of Fish in FBPs detection
Different ways of increasing sensitivity of FISH
Less signal intensity can be identified as a major drawback of traditional FISH. Some researchers stated that FISH is less sensitive with compare to PCR techniques and even convectional culturing methods.3,42 Less permeabilization of target nucleotides and less nucleotide density of slowly growing, very small or metabolically inactive cells can largely accountable for signal depletion. Yet, permeabilisation should carefully balance to protect cell integrity.12 Therefore; an array of improved techniques is introduced to amplify the fluorescence signals in different steps of FISH.
Increase sensitivity in sample preparation step
This step basically comprises of pre-enrichment and microbial separation techniques, sometimes exploiting food matrix and species specific approaches.
Pre-enrichment
Culture enrichment can perform even before FISH, which reported a considerable enhancement of detection sensitivity of FBPs.29,42 This enables multiplication of cells and nucleotides. Higher the targets, brighter the fluorescence signals. Enrichment period vary with the species. For instance, E-coli, an organism with high ribosomal content required comparatively less pre-enrichment time than Camphylobactor spp. or Listaria spp. Dilution of the interfering food matrix components is an additional advantage of enrichment. However, this step largely prolongs the detection time and do not improve the detection of viable but non culturable bacteria. Further, this method restricted the accurate determination and quantification of initial microbial load.4
Microbial separation techniques
Centrifugation (for liquid food matrices); mechanical, chemical or enzymatic separation (for solid food matrices) and filtration are the simplest, target non-specific isolation techniques. Sometimes interfering materials also isolated alone with microbes.4 Further, centrifugation can adversely affect the pathogen viability as the mechanical pressure developed and heat generated during the process. Therefore, pathogen specific isolation techniques are required. Immunogenic separation techniques (IMS) which use a bead coated with pathogen specific antibody43 and magnetic separation techniques (MS) which use a bead coated with metal hydroxides or lectins are examples for such methods.13 Tape FISH, Section FISH, and FISH on the filter are some other techniques developed to separate a representative sample for FISH analysis, also utilized in FBP
detection. Tape fish acquired in detection of salmonella spp. in tomato (Table 1). Camphylobacter spp. and gallibacterium spp. detection in chicken liver and spleen were accomplished with the aid of section FISH.44,45 FISH on filter also utilized in FBPs detection of large variety of food matrices as milk, smoked salmon, cheese, ham, meat, beef, ikura (Japanese food made from salmon) and cabbage.30,33,34,40 However, the method is most effective in liquid foods.4
In tape FISH, sterile and transparent adhesive tape is attached to the food surface, which has the possibility of pathogens localization. FISH can directly perform on the tape or else the tape can contact with a specific culture media to enrich pathogens and detect micro-colonies via microscope. The efficiency of cell capture/release by tape is depended on cell surface properties (e.g. moisture content, surface coatings) and mode of attachment (nonspecific adhesion/adhesion mediated by a specific structure as flagella). It is reported that 99% capture ability of fungi tape.4,31 These tapes are commercially available under different brand names. Further, untreated food component can dip in specific broth for overnight to separate and enrich cells, a time-consuming approach.46 Thin section of food sample as a tissue is used in section FISH. In FISH in filter method, microorganisms are separated into a filter membrane and FISH is performed on the filter or else enrich the microbes in a specific media. These methods are reduced unwanted inhibitors, hence, advantageous than the homogenizing whole sample.4
Increase sensitivity in hybridization step
This aspect is achieved through alteration of probe or target or else hybridization conditions as temperature and buffering capacity. This is the maximum diversified step in the FISH protocol. Yet, each and every diversification was not exploited in FBPs detection except PNA or LNA probing, multicolor FISH and flow FISH.
Multiple probes for one target microorganism
More than one probe can use to target multiple sites of 16S or 23S rRNA of target pathogen. Signals are amplified in 2-3 fold compared to traditional FISH. Normally, it is difficult to design several probes with the same specificity.12
Multiple targets to a probe
Target oligonucleotides can increase artificially. For instance, a sample is incubated with chloramphenicol (CM), an inhibitor of protein synthesis, rRNA degradation and cell division, thus leading to an accumulation of rRNA in the cell. It is testified that, CM treatment increased the percentage of fluorescent cells to nearly 100% compared to untreated cells.47 Antibiotic activity of CM can adversely affect to microbial population, is a limitation of this method.4
Helper oligonucleotides
They are unlabeled nucleotides, are attached to the surrounding of the target site, hence open and facilitate the binding towards the target. This reported 25 fold signal amplification.4
PNA probes
PNA is a synthesized DNA in which the sugar phosphate backbone (negative charge) is replaced with a pseudo-peptide having comparatively high affinity to target. Hence, length of the probe can be shortened. Further, PNA can bind to the structures which DNA cannot. For instance, E. coli O157 serotype was specifically identified by PNA, which DNA could not.25 It is stated that PNA can hybridize in low salt concentration and high temperatures, whereas secondary structures consisting out of DNA or RNA dissolve.4 Further, unipolar nature of PNA increase the cell permeabilization, hence, reduces the hybridization time while enhancing the efficiency. PNA-FISH has reported 5 fold signal amplification than DNA.12 Comparatively high cost and lesser predictability of the results have avoided the application of PNA. However, the use of PNA probes in FBPs detection has increased significantly in recent years, particularly in dairy and poultry products.25,27,41,48,49
Locked nucleic acids (LNA)
This is a RNA analog, which the ribose ring locked by a methylene linkage between the 2′-oxygen and the 4′-carbon. This reduces the conformational flexibility of the ribose and increasing the local organization of the phosphate backbone. Enhancement of melting temperature is the basic advantage of LNA which can speed the hybridization process.50 LNA was least utilize in FBPs identification with compare to PNA. However, Rohde et al. initiated to detect Yersinia spp. in pork using LNA probe in 2017.24 PNA and LNA are unnatural molecules, and they are not substrates for enzymes that modify DNA, RNA and proteins. Therefore, much more effective than DNA or RNA in FISH detection.4
Bacterial chromosomal painting (BCP)
The whole genome of the target organism is used as a probe. In here, all 24 chromosomes can be detected by allocating different color for each. Prolonging the hybridization time is a drawback of this method. BCP has been shown to permit differentiation of Salmonella serotypes51 and has also been applied to marine samples.52 This is the principle behind multicolor FISH (mFISH) and spectral cariotype (SKY). Salmonella spp., Listeria monocytogenes, Enterobacteriaceae and Pseudomonas spp. of bovine and sheep milk were tested using mFISH.26,53
Catalyzed reporter deposition–CARD-FISH
This is an enzyme-mediated signal amplification method, also called TSA (tyramide signal amplification) FISH. In here, horseradish peroxidase (HRP)-labeled oligonucleotide probes are utilized. In the presence of hydrogen peroxide, HRP converts tyramide (amine) into a radical intermediate which has the ability to bind to the electron-rich surfaces of target oligonucleotides. Requirement of vigorous pre-treatments (e.g. proteinase/lysozyme application enables the HRP to enter the cell) and hence loss of cell integrity is a drawback of this method. The method is heavily practice in marine biology.54–56
Polynucleotide probes and RING-FISH
Polynucleotide probes can range in length between 100 and several hundred base pairs. They are made of ssRNA or dsDNA, and carry multiple labels, either fluorescent dyes or digoxigenin/biotin for a secondary detection. This long probes are creating secondary network structure in intracellular and extracellular level, which allows ring shape fluorescence signals.57,58 Due to its high copy number rRNA considered as the only suitable target for FISH. Instead of that targeting other nucleotides as mRNA, tmRNA, plasmid and chromosomal DNA also facilitate by RING-FISH and CARD-FISH. Numerous other FISH techniques were discussed by different authors particularly in medical diagnostic and environmental biological fields as fiber FISH, dope-FISH, raman- FISH and so on (Volpi and Bridger, 2008) which was not or rarely apply in FBPs detection up to date.
Increase sensitivity in visualization step
Fluoresce microscopy is the convectional FISH visualization technique which is surpassing by flow cytometer (Figure 1) due to its unique advantages as not rely on slides, manual counting and large sample can detect rapidly without labor cost.4
Flow cytometry (flow-FISH)
In flow FISH, laser-based technology is applied to count, sort, and profile cells in a heterogeneous fluid mixture. Therefore, the foods as tomato, spinach, spices and particularly milk are tested for FBPs using flow FISH.4,31,35,41
Bacterial chromosomal painting (BCP)
The whole genome of the target organism is used as a probe. In here, all 24 chromosomes can be detected by allocating different color for each. Prolonging the hybridization time is a drawback of this method. BCP has been shown to permit differentiation of Salmonella serotypes51 and has also been applied to marine samples.52 This is the principle behind multicolor FISH (mFISH) and spectral cariotype (SKY). Salmonella spp., Listeria monocytogenes, Enterobacteriaceae and Pseudomonas spp. of bovine and sheep milk were tested using mFISH.26,53
Catalyzed reporter deposition–CARD-FISH
This is an enzyme-mediated signal amplification method, also called TSA (tyramide signal amplification) FISH. In here, horseradish peroxidase (HRP)-labeled oligonucleotide probes are utilized. In the presence of hydrogen peroxide, HRP converts tyramide (amine) into a radical intermediate which has the ability to bind to the electron-rich surfaces of target oligonucleotides. Requirement of vigorous pre-treatments (e.g. proteinase/lysozyme application enables the HRP to enter the cell) and hence loss of cell integrity is a drawback of this method. The method is heavily practice in marine biology.54–56
Polynucleotide probes and RING-FISH
Polynucleotide probes can range in length between 100 and several hundred base pairs. They are made of ssRNA or dsDNA, and carry multiple labels, either fluorescent dyes or digoxigenin/biotin for a secondary detection. This long probes are creating secondary network structure in intracellular and extracellular level, which allows ring shape fluorescence signals.57,58 Due to its high copy number rRNA considered as the only suitable target for FISH. Instead of that targeting other nucleotides as mRNA, tmRNA, plasmid and chromosomal DNA also facilitate by RING-FISH and CARD-FISH. Numerous other FISH techniques were discussed by different authors particularly in medical diagnostic and environmental biological fields as fiber FISH, dope-FISH, raman- FISH and so on (Volpi and Bridger, 2008) which was not or rarely apply in FBPs detection up to date.
Increase sensitivity in visualization step
Fluoresce microscopy is the convectional FISH visualization technique which is surpassing by flow cytometer (Figure 1) due to its unique advantages as not rely on slides, manual counting and large sample can detect rapidly without labor cost.4
Flow cytometry (flow-FISH)
In flow FISH, laser-based technology is applied to count, sort, and profile cells in a heterogeneous fluid mixture. Therefore, the foods as tomato, spinach, spices and particularly milk are tested for FBPs using flow FISH.4,31,35,41
FISH is an immerging trend in FBPs detection which is in its basic stage with compare to PCR based detection techniques. Some FISH protocols acquire as high as 85-90% of its total time for pre-enrichment step.40 Therefore, if scientists can find solutions to omit or automate the pre-enrichment step, quick results can obtain along with high sensitivity. Available literature demonstrated that FISH can successfully exploit in FBPs detection. However, adaptation of alterations to enhance the sensitivity is must to ensure great accuracy and sensitivity. Lack of standard protocols and automation steps, investigator has to anticipate the potential pathogen present in the food sample when selecting a probe are also drawbacks in FISH. Each and every FBPs detection method has pros and cons. Hence, selecting a suitable method is depending on objective, available facilities and finally the impact of the finding to the general population. For instance, developing countries are still relying on convectional culture base techniques and expired technologies can be a possible cause for their elevating counts of FBP outbreaks.
None.
The author declares no conflict of interest.

©2018 Dias, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.