International Journal of
eISSN: 2573-2889


Research Article Volume 4 Issue 6
1Department of Biochemistry and Cell Biology, University of Abomey-Calavi, Benin
2Laboratory of Research in Applied Biology, Polytechnic School of Abomey-Calavi, University of Abomey-Calavi, Benin
Correspondence: Nicodème Worou Chabi, Laboratory of Biochemistry and Molecular Biology, Department of Biochemistry and Cell Biology, Faculty of Science and Technology, University of Abomey-Calavi, Benin, Tel +22997084906,
Received: November 19, 2019 | Published: December 9, 2019
Citation: Chabi NW, Sognigbé BG, Duéguénon E, et al. Evaluation of the genetic susceptibility to the metabolic syndrome by the CAPN10 SNP19 gene in the population of South Benin. Int J Mol Biol Open Access. 2019;4(6):196-200. DOI: 10.15406/ijmboa.2019.04.00120
Metabolic syndrome is a multifactorial disorder whose etiology is resulting from the interaction between genetic and environmental factors. Calpain 10 (CAPN10) is the first gene associated with type 2 diabetes that has been identified by positional cloning with sequencing method. This gene codes for cysteine protease; ubiquitously expressed in all tissues, it is involved in the fundamental physiopathological aspects of insulin resistance and insulin secretion of type 2 diabetes. The goal of this study was to evaluate the genetic susceptibility to the metabolic syndrome by the CAPN10 gene in the population of southern Benin. This study involved apparently healthy individuals’ aged 18 to 80 in four ethnic groups in southern Benin. It included 74 subjects with metabolic syndrome and 323 non-metabolic syndrome patients who served as controls, with 222 women versus 175 men with an average age of 40.58 ± 14.03 years old. All subjects were genotyped for the SNP 19 polymorphism of the CAPN10 gene with the PCR method in order to find associations between this polymorphism and the metabolic syndrome. We found an association between this polymorphism and a risk of developing the metabolic syndrome only in the Tori and Nago ethnic groups. Genotype 1/1, a risk factor for the occurrence of the metabolic syndrome, is associated with environmental and nutritional factors in the four ethnic groups studied. The results of our study show that only the Goun and Nago ethnic groups have a genetic predisposition to the metabolic syndrome toward CAPN10 gene.
Keywords: metabolic syndrome, capn10 gene, type 2 diabetes, ethnic groups, Southern Benin
The metabolic syndrome is the coexistence of a set of metabolic aberrations, mainly, insulin resistance, obesity (general and abdominal), dysglycemia, dyslipidemia and high blood pressure.1 Its prevalence is 17.1% in southern Benin. Environmental factors such as ethnic group, age, sex, educational level, marital status, physical inactivity, overweight, non-regular consumption of fruits and vegetables and high cholesterol LDL are critical determinants of metabolic syndrome. 2 In addition, genetic factors have contributed to individual susceptibility to metabolic syndrome.3,4
Many works have studied the associations between genetic polymorphisms and the various components of the metabolic syndrome. Very few, on the other hand, have explored this association with the metabolic syndrome as an entity, that is, according to the criteria of the International Diabetes Federation.5 These studies have shown that variations in the calpain 10 gene (CAPN10) are significantly associated with the metabolic syndrome.6 The Calpain 10 is the first gene with 2-of-the-Genome-Human-Key-to-Buy with Positional Cloning and Sequencing. Located on the short arm of chromosome 2 (2p.37.3), this gene codes for one over the protein of proteases;7 ubiquitously expressed in all tissues, in the fundamental pathophysiological aspects of insulin resistance and insulin secretion of type 2 diabetes.8 In addition, the CAPN10 gene is associated with cholesterol levels and blood pressure.6
A recent study has shown an association between the SNP19 polymorphism of the calpain 10 gene with type 2 diabetes in an ethnic group in the Tunisian populations. Allele 2 has been reported to provide protection against the onset of type 2 diabetes, but Allele 1 is a risk factor and should be considered in the assessment of a predisposition to such conditions. as diabetes, obesity, metabolic syndrome diseases, high blood pressure and cardiovascular diseases. The objective of this work was to evaluate the genetic susceptibility to the metabolic syndrome by the CAPN10 gene in the population of southern Benin.
Topics analyzed
The participants were recruited as part of the study on the metabolic syndrome in the three communes of southern Benin in 2018: the commune of Avrankou in the department of Ouémé; the communes of Ifangni and Sakété in the plateau department. The sample includes individuals aged 18 to 80 randomly selected from four ethnic groups. The first condition for inclusion in this study is to be an adult and to have participated in the two previous studies.2 The second is to belong to one of the Goun, Tori, Nago and Yoruba ethnic groups. The third condition is to have accepted to participate in the study and to have given written consent. The subjects are distributed as follows: 74 with the metabolic syndrome and 360 healthy subjects. Subjects with the metabolic syndrome were detected based on criteria from the International Diabetes Federation. The characteristics of the study population are summarized in Tables 1&2.
Parameters |
Metabolic syndrome |
|
|
Presence (n/%) |
Absence (n/%) |
p-value |
|
Age ˂ 40 |
18 (7.8) |
212 (92.2) |
0 |
Age ≥ 40 |
56 (27.5) |
148 (72.5) |
|
Female |
61 (25.7) |
176 (74.3) |
0 |
Male |
13 (6.6) |
184 (93.4) |
|
Educated |
23 (10.5) |
196 (89.5) |
0 |
Not educated |
51 (23.7) |
164 (76.3) |
|
Married |
67 (19.1) |
284 (80.9) |
0.02 |
Not married |
7 (8.4) |
76 (91.6) |
|
History of HTA |
10 (14.5) |
59 (85.5) |
0.537 |
No history of hypertension |
64 (17.5) |
301 (82.5) |
|
History of diabetes |
1 (7.7) |
12 (92.3) |
0.362 |
No history of diabetes |
73 (17.3) |
348 (82.7) |
|
Regular physical activity |
1 (2.3) |
42 (97.7) |
0.006 |
No regular physical activity |
73 (18.7) |
31 (81.3) |
|
IMC ˂ 25 kg/m2 |
20 (6.9) |
268 (93.1) |
0 |
IMC ≥ 25 kg/m2 |
54 (37.0) |
92 (63.0) |
|
The data is expressed in n (%); HTA = Hypertension; BMI = Body Mass Index.
Table 1 Distribution of metabolic syndrome according to IDF criteria2
Parameters |
Metabolic syndrome |
|
|
Presence (n/%) |
Absence (n/%) |
P-value |
|
Alcohol consumption |
37 (19.8) |
150 (80.2) |
0.187 |
No alcohol consumption |
37 (15.0) |
210 (85.0) |
|
Tobacco consumption |
5 (19.2) |
21 (80.8) |
0.76 |
No smoking |
69 (16.9) |
33 (83.1) |
|
Regular consumption of fruits and vegetables |
2 (4.6) |
41 (95.4) |
0.022 |
No regular consumption of fruits and vegetables |
72 (18.4) |
319 (81.6) |
|
Total hypercholesterolemia |
15 (24.2) |
47 (75.8) |
0.106 |
No total hypercholesterolemia |
59 (15.9) |
313 (84.1) |
|
LDL hypercholesterolemia |
41 (27.5) |
108 (72.5) |
0 |
No LDL hypercholesterolemia |
33 (11.6) |
252 (88.4) |
|
Data are expressed in n (%)
Table 2 Nutritional factors associated with metabolic syndrome2
Data gathering
This study uses data from previous studies, including data on age, weight, height, sex, regular consumption of fruits and vegetables, lifestyle, level of education, diet, socio-economic conditions, physical activity and biochemical parameters.2
Methods
DNA extraction: The genomic DNA of participants with or without metabolic syndrome of the four ethnic groups was extracted from leukocytes from 10 ml of EDTA whole blood, using a DNA isolation kit (Kit Nucleon Bac Pharmacia-Biotch from Amersham Company) in the Laboratory of Biochemistry and Molecular Biology located at the Institute of Applied Biomedical Sciences (ISBA) in Cotonou, Benin. The optical density was taken using a Nanodrop Lite 8000 spectrophotometer from Thermo Scientific.
Genotyping: Genotyping of the UCSNP19 polymorphism of the calpain gene consists of an insertion/deletion characterized by a two or three-fold repetition of a 32 bp sequence.7 The detection of the UCSNP19 mutation of the calpain 10 gene was carried out by PCR amplification according to the protocol described by Baroudi using the following primers: F: 5'-GTT TGG TTC TCT TCA GCG TGG AG - 3'and R: 5'-CAT GAA CCC TGG CAG GGT CTA AG-3. The genomic DNA (50 ng) was amplified in a final volume of 25 .mu.l, containing 1μmol/L of each primer, 1 mM of dNTP, 1.5 mol/L of MgCl2, 5μL of the 5X PCR buffer and 0.25U of the GoTaq DNA polymerase (Promega). The amplification conditions were as follows: an initial denaturation at 95°C for 5 minutes, 30 PCR cycles each comprising denaturation at 94°C for 60sec, hybridization at 55°C for 60sec, extension at 72°C for 60 sec and finally a final elongation at 72°C for 10 minutes.
The PCR products were separated by agarose gel electrophoresis at 2% gel stained with 5 μg/ml red gel. A 100 bp DNA molecular weight marker (ladder) was used to compare the size of amplification fragments. The migration was performed at a scale of 80 V/cm for 30 min. The revelation was made by ultraviolet light. The presence of the wild-type allele 1 results in the visualization of a 155 bp band (deletion of the 32 bp repeat) and that of the allele 2 by that of a 187 bp band (insertion of the repetition 32 bp).
Statistical analysis
The information collected was analyzed using Excel and R version 3.2.2 with the FactoMineR package. Quantitative data were presented by averages and standard deviation (SD) and qualitative data by their size (n) and proportion (%). The deviation of the Hardy-Weinberg equilibrium was tested using the χ2 (1 df) test. The statistical calculation is based on the use of Student's t-test. Allelic and genotypic frequencies were compared by a χ2 test. The relative risk associated with the genotype was estimated by calculating the odds ratio (OR) at 95% [CI]. We assessed the association between CAPN10 genotypes and risk factors associated with metabolic syndrome using logistic regression (OR and 95% [CI]). The 0.05 threshold was used for statistical significance.
Sociodemographic characteristics of the study population
A total of 397 subjects were included in this study in both departments. Among them 175 (44.08%) were male and 222 (55.92%) female with a sex ratio of 0.79.
The mean age of men was 38.89±12.78 compared to 41.90±14.83 for women. The average age of the participants was 40.58±14.03 with extremes of 18 to 80 years (Table 3).
Parameters |
Male |
Female |
Total |
n= 175 |
n= 222 |
n= 397 |
|
Average age (years) |
38,89±12,78 |
41.90±14,83 |
40,58±14,03 |
level of education |
|||
No |
53 (25.98) |
151 (74.02) |
204 (51.39) |
Primary |
46 (59.74) |
31 (40.26) |
77 (19.40) |
Secondary |
54 (58.70) |
38 (41.30) |
92 (23.17) |
Superior |
22 (91.67) |
2 (8.33) |
24 (6.05) |
Marital status |
|||
Single |
31 (67.39) |
15 (32.61) |
46 (11.59) |
Married |
142 (44.24) |
179 (55.76) |
321 (80.86) |
Divorced or widowed |
2 (6.67) |
28 (93.33) |
30 (7.56) |
Ethnic group |
|||
Goun |
56 (45.16) |
68 (54.84) |
124 (31.23) |
Tori |
60 (46.88) |
68 (53.13) |
130 (32.75) |
Nago |
44 (41.90) |
61 (58.10) |
103 (25.94) |
Yoruba |
15 (37.50) |
25 (62.50) |
40 (10.08) |
Table 3 Sociodemographic characteristics of the study population
Data expressed as mean ±SD or frequency (%)
Clinical characteristics of the study population
Table 4 presents the clinical characteristics of subjects with metabolic syndrome as well as apparently healthy subjects. From the analysis of the table, it appears that from clinical parameters, there is a statistically significant difference between the two groups of subjects.
Metabolic syndrome |
|||
|
Presence |
Absence |
p-value |
Parameters |
|||
Total number (F / M) |
74 (61/13) |
323 (161/162) |
3.526E-07* |
Average age (years) |
48,31±12,89 |
38,80±13,69 |
1.18E-07* |
Weight in kg |
74,00±15,27 |
62,48±12,19 |
2.66E-08* |
BMI (kg / m2) |
28,21±5,41 |
22,97±4,01 |
6.66E-12* |
TT in cm |
93,88±10,89 |
80,98±10,07 |
2.39E-15* |
NOT in mmHg |
147,23±22,20 |
123,66±19,67 |
2.86E-13* |
PAD in mmHg |
91,85 ±12,37 |
78,15±12,94 |
8.08E-14* |
Glycemia in g / l |
1,00±0,27 |
0,81±0,16 |
1.06E-08* |
Triglycerides in g / l |
1,14±0,51 |
0,85±0,31 |
1.10E-05* |
HDL-cholesterol in g / l |
0,43±0,11 |
0,55±0,17 |
5.21E-13* |
Data expressed as mean ±SD or frequency (%); * statistically significant
Table 4 Clinical characteristics of subjects with metabolic syndrome and healthy subjects
The different fragments observed with the UV lamp
The gel photograph identified the different genotypes in the study population (Figure 1)
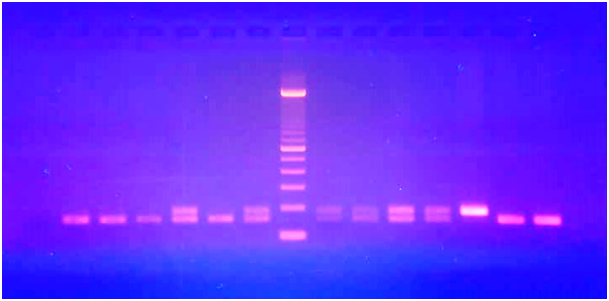
M: molecular weight marker, TN: negative control, 1/1: 155 bp, 1/2: 155 and 187, 2/2: 187bp
Figure 1 Migration photo of fragments CAPN10 gene amplification.
Genotypic and allelic distribution of the SNP 19 mutation of the CAPN 10 gene in the study population
The analysis of the UCSNP19 polymorphism of the CAPN 10 gene allowed us to estimate the frequency of the allele 2 in the population with the metabolic syndrome where it was equal to 16.7%, 34.2%, 23.8% and 46.2% respectively in the Goun, Tori, Nago and Yoruba. In the apparently healthy individuals of the Goun, Tori, Nago and Yoruba ethnic groups, this frequency is respectively 40.3%, 38.3%, 59.9% and 40.7%. In the Tori and Yoruba ethnic groups, the genotypic distribution is consistent with Hardy Weinberg's equilibrium. On the other hand, at Goun and Nago we notice a deviation of Hardy Weinberg's equilibrium (Table 5).
|
Genotype (%) |
|
Allèle (%) |
|
|
|||
|
1//1* |
1//2 |
2//2 |
P |
1* |
2 |
OR (95% CI) |
P |
Study population |
||||||||
SMet presence (n = 74) |
43(58.1) |
20(27.0) |
11(14.9) |
0.008* |
106(71.6) |
42(28.4) |
1.96 (1.31-2.97) |
0.001* |
Absence SMet (n = 323) |
127(39.3) |
107(33.1) |
89 (27.6) |
360(56.2) |
280(43.8) |
|||
GOUN |
||||||||
SMet presence (n = 21) |
16(76.2) |
3(14.3) |
2(9.5) |
0.009* |
35(83.3) |
7(16.7) |
3.36 (1.38-9.40) |
0.004* |
Absence SMet (n = 103) |
41(39.8) |
41(39.8) |
21(20.4) |
123(59.7) |
83(40.3) |
|||
TORI |
||||||||
SMet presence (n = 19) |
9(47.4) |
7(36.8) |
3(15.8) |
0.883 |
25(65.8) |
13(34.2) |
1.19 (0.55-2.68) |
0.718 |
Absence SMet (n = 111) |
49(44.1) |
39(35.1) |
23(20.7) |
137(61.7) |
85(38.3) |
|||
NAGO |
||||||||
SMet presence (n = 21) |
14(66.7) |
4(19.0) |
3(14.3) |
0.006* |
32(76.2) |
10(23.8) |
4.37 (1.94-10.66) |
0.000* |
Absence SMet (n = 82) |
25(30.5) |
19(23.2) |
38(46.3) |
69(42.1) |
95(57.9) |
|||
YORUBA |
||||||||
SMet presence (n = 13) |
4(30.8) |
6(46.2) |
3(23.1) |
0.57 |
14(53.8) |
12(46.2) |
||
Absence SMet (n = 27) |
12(44.4) |
8(29.6) |
7(25.9) |
|
32(59.3) |
22(40.7) |
0.80 (0.28-2.30) |
0.809 |
* Statistiquement significant
Table 5 Genotypic and allelic frequencies of the SNP 19 mutation of the CAPN10 gene in the population of southern Benin
The same table shows that in Goun metabolic syndrome carriers, 16 (76.2%) were homozygous 1/1, 3 (14.3%) were heterozygous 1/2 and 2 (9.5%) were homozygous 2/2. In Tori metabolic syndrome carriers, 9 (47.4%) were homozygous 1/1, 7 (36.8%) were heterozygous 1/2 and 3 subjects (15.8%) were homozygous 2/2. In Nago metabolic syndrome carriers, 14 (66.7%) were homozygous 1/1, 4 (19.0%) were heterozygous 1/2 and 3(14.3%) were homozygous 2/2. In Yoruba metabolic syndrome carriers, 4 (30.8%) were homozygous 1/1, 6 (46.2%) were heterozygous 1/2 and 3 (23.1%) were homozygous 2/2. The table does not show a statistically significant difference in the genotypic distribution between the Tori and Yoruba ethnic groups (p=0.883 and p=0.57 respectively). On the other hand, there is a statistically significant difference in the genotypic distribution between the Goun and Nago ethnic groups (p=0.009 and p=0.006 respectively). The statistical analysis performed in the four ethnic groups showed a different association between the studied marker and the susceptibility to the metabolic syndrome. Indeed, the study of the allelic and genotypic distribution shows only in Goun and Nago that the allele 2 is a protective factor; on the other hand, the allele 1 seems to be a risk factor for the metabolic syndrome, [p=0.004; OR=3.36 (1.38-9.40) and p=0.000; OR=4.37 (1.94-10.36) respectively]. Moreover, in Tori and Yoruba, no significant association was observed between the carriers of metabolic syndrome and the apparently healthy individuals allowed to define the polymorphism of CAPN 10 as a neutral marker with metabolic syndrome (Table 5).
Multivariate logistic regression analysis did not show a difference in genotype distribution 1/1 by age, educational level, marital status and regular consumption of fruits and vegetables (p>0.05). In contrast, a statistically significant difference was found by sex, physical activity, BMI, blood glucose, HDL cholesterol and triglyceride levels (p<0.05) (Table 6).
|
ß |
SE |
P |
SMet versus healthy |
|||
Age |
-0.011 |
0.019 |
0.567 |
Sex |
-4.084 |
0.85 |
1.55e-06 * |
Level of education |
-0.828 |
0.622 |
0.183 |
Marital status |
0.008 |
0.035 |
0.8 |
Physical activity |
0.327 |
1.487 |
0.002396 * |
Cons fruit |
0.468 |
0.698 |
0.502 |
BMI |
-0.057 |
0.098 |
0.000279 * |
Blood Sugar |
-0.057 |
0.018 |
0.002396 * |
Chol HDL |
14.154 |
2.431 |
5.87e-09 * |
Triglycerides |
-3.194 |
0.715 |
8.09e-06 * |
* Statistically significant
Table 6 Logistic regression by metabolic syndrome as dependent variable
The present study is, to our knowledge, the first study carried out in Benin in the exploration of the genetic susceptibility to the metabolic syndrome in a population in apparent health. It allowed to study the UCSNP19 polymorphism of the CAPN10 gene in relation to environmental and nutritional factors in a population of 397 subjects with or without metabolic syndrome. To achieve this, we used a candidate gene approach and conducted an association study between the UCSNP19 polymorphism and the risk of metabolic syndrome3,9 in four (4) ethnic groups in southern Benin. We looked at the CAPN10 gene encoding cysteine protease because, based on literature data, it is involved in the basic physiopathological aspects of insulin resistance and insulin secretion of type 2 diabetes.8 It is therefore a good gene candidate relevant for the evaluation of a genetic predisposition to the occurrence of the metabolic syndrome. The study population was young, consisting of 397 individuals with an average age of 40.58 ± 14.03. This sample size is fairly representative of the southern Benin population for evaluating allelic and genotypic frequencies. Our sample is similar in size to that of a study in Polish European populations,10 but it is higher than that of Baroudi.
The statistical analyzes performed in the four ethnic groups show different associations between the studied marker and the genetic susceptibility to the metabolic syndrome. In fact, allelic and genotypic distributions allowed to define the risk factors only in Goun and Nago, in which allele 1 constitutes a risk factor [p=0.004; OR=3.36 (1.38-9.40) and p=0.000; OR=4.37 (1.94-10.36)]; however, the allele 2 seems to be protective against the metabolic syndrome. In the Tori and Yoruba ethnic groups, there is not a significant difference between metabolic syndrome carriers and apparently healthy subjects for defining SNP19 polymorphism as a neutral marker for metabolic syndrome. This study confirms the difference between ethnic groups in the genetic predisposition to metabolic syndrome as reported in the literature.11
One of the explanatory factors for this difference is the environment. Indeed, in these four ethnic groups of the population of southern Benin, there is a disparity (p=0.009) in the distribution of BMI to obesity (BMI ≥ 30 Kg/m2).2 The heterogeneity of the distribution of the SNP19 polymorphism of the CAPN 10 gene is multifactorial and illustrates well the gene-environment interactions. Indeed, we also analyzed the interaction of environmental parameters (physical activity, educational level, marital status, and BMI) and nutritional (regular consumption of fruits and vegetables, blood glucose, HDL cholesterol and triglycerides) with genotype 1/1 (Table 6). In our study population, we found an association between genotype 1/1 and sex, physical activity, BMI, blood glucose, HDL cholesterol and triglyceride. The results of this study suggest that environmental and nutritional factors greatly affect population health. Indeed, the four ethnic groups have almost the same eating habits composed largely of oil and starchy foods.2 This type of diet favors the metabolic syndrome.12–14
The results of our study show that only the Goun and Nago ethnic groups have a genetic predisposition to metabolic syndrome with SNP19 of CAPN10 gene. Genotype 1/1, a risk factor for the occurrence of metabolic syndrome, is associated with environmental and nutritional factors in the four ethnic groups studied.
None.
The author declares there is no conflicts of interest.

©2019 Chabi, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.