International Journal of
eISSN: 2573-2889


Mini Review Volume 7 Issue 1
Instituto Politécnico Nacional, Laboratorio de Biotecnología Molecular, México
Correspondence: Sehom Rivera Gutiérrez, Instituto Politécnico Nacional, Laboratorio de Biotecnología Molecular, Unidad Profesional Interdisciplinaria de Biotecnología, México
Received: March 15, 2024 | Published: March 22, 2024
Citation: Gutiérrez SR, Durán NV. Biogenesis and function of microRNAS from plants, animals and fungi applicable in health and food biotechnology. Int J Mol Biol Open Access. 2024;7(1):41-47. DOI: 10.15406/ijmboa.2024.07.00166
MicroRNAs are small non-coding RNAs found in plants, animals and fungi. They are involved in developmental, metabolic, growth and reproduction processes. They modulate these processes by regulating target mRNAs by binding to them through base complementarity, and with the help of an enzymatic complex, they cleave the mRNA, or interrupt its translation into protein. Its biogenesis in plants occurs within the nucleus, to later be exported to the cytoplasm where it will take its regulatory action. In animals, biogenesis begins in the nucleus, but unlike plants, it is completed in the cytoplasm. A biosynthetic mechanism has not been established in fungi, although it could be similar to that in plants due to the nature of the fungal transcripts. MicroRNAs can be used as key biomolecules in the solution of health problems, in the improvement of plants of agronomic interest and in the optimization of bioprocesses to obtain industrial products of food interest. Furthermore, they have been proposed as strategic biomolecules in the design of nutritional tables that offer a better quality of life to people. Agronomically, its study focuses mainly on improving the characteristics of crops of commercial interest, as well as combating pests that affect the production of fruits, vegetables and grains. For this reason, microRNAs are being proposed as cutting-edge molecules for solving health food problems worldwide.
Keywords: biogenesis of microRNAs, animal miRNAs, plant miRNAs, fungal miRNAs, biotechnological applications
MicroRNAs (miRNAs) are small RNAs of 18-25 nucleotides in length1 that regulate gene expression in plants, animals and fungi.2,3 Its mechanism of action is due to sequence complementarity with a target messenger RNA (mRNA);4,5 In general, they repress gene expression by inducing mRNA cleavage or inhibiting its translation6,7 (Figure 1). miRNAs are master regulators of gene expression at the post-transcriptional level,8 and are involved in a diversity of biological processes9 in multiple organisms, such as development and responses to biotic and abiotic stimuli that affect living beings,10 cell proliferation, differentiation and death,1 pathological processes such as cancer or autoimmune diseases, cardiovascular and metabolic diseases,11 in the response to the attack of human pathogens such as Leishmania, Trypanosoma, Toxoplasma and Plasmodium,12 in root formation,13 morphogenesis of the leaves and flowering of plants.14 They can be used as biomarkers to detect15,16 and combat diseases,17 used as biotechnological tools in plants,18 and in obtaining secondary metabolites of commercial interest such as phenols, alkaloids, isoprenoids, glycosides, tannins and triterpenes for the dyes, flavorings, nutraceuticals and insecticides industry.19 It has been estimated that, at least in plants, miRNA-mediated regulation of gene expression dates back 400 million years,20,21 highlighting how evolutionarily conserved they are, although there are also species-specific miRNAs. The biogenesis of miRNAs from animals, plants and fungi has points in common: an RNA polymerase recognizes a segment of the genome where the MIR gene is housed which codes for a miRNA22 and transcribes a primary RNA, which undergoes a self-folding process to form a double-stranded RNA of the stem-loop type,23 which will subsequently be cut by an enzymatic microprocessor at both ends of its structure, generating an RNA of ~22 nt; said RNA will be loaded into the Argonaute (AGO) proteins to form an RNA-Induced Silencing Complex (miRISC),24 where the AGO proteins will be responsible for directing gene repression This review presents the main similarities and differences in the function of miRNAs from plants, animals and fungi, focusing mainly on the mechanisms of biogenesis and postulating that miRNAs are not only master regulators of gene expression, but also as a tool cutting-edge for biotechnological applications such as the genetic improvement of crops and increased yields, their use as molecular biomarkers in the detection and solution of health problems, and their use in obtaining metabolites of industrial interest.
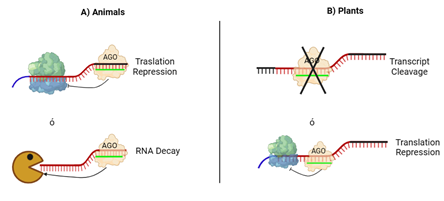
Figure 1 Mechanism of action of miRNAs.
(A) In the case of miRNAs of animal origin, the miRNA is linked by base complementarity to an mRNA, preventing the ribosome from translating the protein (top), or in some cases, the mRNA is degraded to prevent the translation of the messenger (bottom-packman).
(B) in the case of plant miRNAs, generally the miRNA binds by total base complementarity to an mRNA, which causes the protein Argonaute (AGO) to cleave the mRNA, preventing translation into protein (top), but in sometimes, the complementarity is not complete and the mRNA is not translated into protein, although this complementarity is sufficient to prevent the ribosome from carrying out the translation (bottom).
Biogenesis and functon of microRNAs in animals
In 1993, the first miRNA was discovered in Caenorhabditis elegans, miRNA lin-4,25 and in 2000 in that same organism, the second miRNA, let-7, was discovered.26 These discoveries prompted the study of miRNAs in many working groups around the world, and thanks to the large amount of information obtained, it has been estimated that these molecules control around two-thirds of all mammalian coding genes.27 In animals, the biosynthesis of miRNAs begins in the nucleus of cells (Figure 2) where RNA polymerase II (Pol II) transcribes the respective MIR genes to obtain transcripts of 300-1000 nucleotides in length that self-fold by taking the stem-loop form, which are called primary miRNAs (pri-miRNAs).28–30 For Pol II to bind more strongly to the promoter region of the miRNA, it must be dephosphorylated, and once bound, Pol II must be phosphorylated at the C-terminal end of the large subunit to switch from initiation mode to elongation mode of the growing transcript,29 thus obtaining the stem-loop of pri-miRNA, which is methylguanylated at the 5' end and polyadenylated at the 3' end.31 At that moment, the pri-miRNA is recognized by Drosha-like type III RNase and by DGR8,32,33 thus forming a processing complex that will cleave the 5' and 3' ends of the pri-miRNA within the cell nucleus, to yield a precursor miRNA (pre-miRNA) of 70-90 nt in length.28,29,34 Under these conditions, the pre-miRNA is exported to the cytoplasm by Ran-GTP and a member protein of the Ran-dependent nuclear transport receptor family, Exportin-5 (Exp5).35 It has been observed that, in vitro, Exp5 binds to pre-miR-30 but only in the presence of Ran-GTP, thus both Ran-GTP and Exp5 being present simultaneously; This step of miRNA maturation, in addition to activating export, protects pre-miR-30 from exonucleotide digestion.36 Once in the cytoplasm, the RNase III Dicer and the cofactor TRBP (transactivation-responsive RNA-binding protein) eliminate the loop to generate a duplex of ~22 nt in length,28,29 then the 5p strand (leading strand; the complementary strand is known as the passenger strand, 3p or miRNA*37) of the miRNA duplex is loaded into an RNA-dependent silencing complex (RISC), where the key member is the AGO enzyme.38–40 Finally, the mature miRNA guides the RISC complex towards the target mRNA, where the miRNA is will partially bind by base complementarity to the 3 UTR region of the mRNA, thus interrupting and blocking the translation of the protein28,41,42 (Figure 1A). The participation of miRNAs in animals is essential,43 and as an example it can be mentioned that they are involved in sex specification, as is the case of miRNAs, miR-125a, miR-9, and the let-7 family, which show overexpression in primordial germ cells in Mus musculus domesticus to commit towards a male fate, while overexpression of miR-29b commits primordial germ cells towards a female fate in mice.44,45 Another example is miRNA-122-5p, which is known to stimulate proliferation and DNA synthesis in humans.46 Precisely in humans, another miRNA, miR-140-5p, is involved in skeletal development, and a mutation in the gene that produces it causes skeletal defects such as short stature, brachydactyly, premature degeneration of intervertebral discs and late epiphyseal ossification of the hip and knee.47
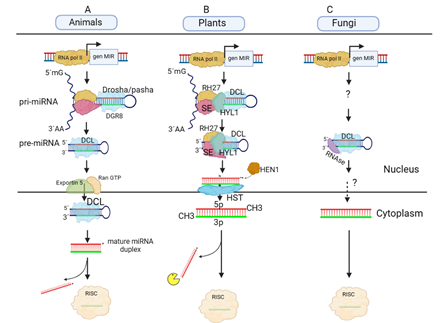
Figure 2 Biogenesis of miRNAs in animals, plants and fungi.
(A) Biogenesis in animals. RNA Pol II transcribes a MIR gene and the transcript self-folds taking the form of a stem-loop called primary miRNA (pri-miRNA). Drosha/pasha recognizes this structure and cleaves the 5' and 3' ends, taking the name precursor miRNA (pre-miRNA). With the help of Ran-GTP, Exportin-5 ejects the pre-miRNA where it is processed by the Dicer protein to yield a mature miRNA duplex. The mature strand is loaded into the RNA-induced silencing complex (RISC), while the complementary strand will take another fate.
(B) Biogenesis in plants. RNA Pol II transcribes a MIR gene and the transcript takes the form of a stem-loop called pri-miRNA, subsequently a microprocessor composed of SERRATE (SE), HIPONASTIC LEAVES 1 (HYL-1), RH27 and Dicer-like-1 (DCL), in a first step, they cleave the 5' and 3' ends to generate the pre-miRNA. In a second step, the microprocessor cleaves the loop to yield an RNA duplex. The duplex is rapidly methylated by the enzyme HUA ENHANCER 1 (HEN1) and at this time, the exportin HASTY ejects the mature duplex into the cytoplasm. Until this moment, the entire maturation and export process occurs simultaneously. Once in the cytoplasm, the duplex strands are separated by the protein Argonaute (AGO) found within the RISC complex, where one strand is loaded into the complex, while the other is generally degraded, or in some cases it is loaded into exosome-like microvesicles to perform other functions.
(C) Biogenesis in fungi. There is no proposed mechanism for the case of fungi, but it is known that polymerase II, Dicer-like and an RNase articipate in the generation of milRNAs (miRNA-like molecules). It is also not known whether milRNA maturation occurs entirely in the nucleus or part of it occurs in the cytoplasm. Image of own creation.
miRNAs in animals were used as molecular biomarkers for the first time in 2008 to detect cancer, diffuse large B cell lymphoma.48 In this same sense, it is known that the levels of mirR-16, miR-22, miR375 and miR-451 are elevated in the serum of patients with Graves' disease,49 and although to date, the specific reason for its increase is not known; these miRNAs can be used as biomarkers in this autoimmune disease. Likewise, they can be used as biomarkers in the detection of tuberculosis since they are involved in the control of Mycobacterium tuberculosis infections by regulating inflammatory responses and the activation of cytokine signaling in humans; some, such as miR-21-5p, are upregulated, while others, such as miR-149, are deregulated in this disease.50 In the case of leishmaniasis, it is known that an infection with L. amazonensis, L. infantum, L. major and L. donovani alters the miRNA profiles of dendritic cells and macrophages; the same behavior occurs in dogs and murine models; Among the miRNAs with an altered profile are miRNA-130a, miRNA-130b, miRNA-125b, miRNA-26a, miRNA-26b, miRna-155, miRNA-21, miRNA-19a, miRNA-23a and miRNA-720.12
In Chagas disease, caused by the parasite Trypanosoma cruzi, an increase in the levels of miR-208a, miR.19a-3p, miR-199b-5p, miR-29b-3p has been observed in the plasma of infected patients,12 and the upregulation of hsa-miR-490-5p, hsa-miR12135 and hsa-miR-210-5p and deregulation of hsa-miR-497-5p, hsa-miR-877-5p and hsa -miR-1271-5p in human placenta explants.51 In a case of toxoplasmosis, a disease caused by Toxoplasma gondii, the miRNA clusters miR-17 ~ miR-72 and miR-106b ~25 are upregulated,52 while in acute ocular toxoplasmosis miR- 155-5p and miR-29c-3p are upregulated while miR-21-5p and miR-125b-5p are deregulated.53 In malaria, a human disease caused by the parasitism of the protozoans Plamodium falciparum, P. vivax, P. ovale, P. malariae and P. knowlesi, the miRNAs miR-451, miR-106, miR-16, miR-92, miR .7b, miR-142, miR-144, let-7f, let-7a and miR-91 are downregulated, while the miRNAs miR-233 and miR19b are upregulated in red blood cells.54
Biogenesis and function of microRNAs in plants
In plants, miRNAs have different functions during plant development,55 they are involved in the post-transcriptional regulation of developmental genes,56,57 and control the production of secondary metabolites such as phenylpropanoids, terpenoids and alkaloids.58 Most miRNAs are encoded by MIR intergenic genes, although some originate from introns of coding or non-coding genes.59,60 As in animals, the biogenesis of miRNAs in plants begins in the nucleus, but in this case, the two miRNA stem-loop cutting steps occur within the nucleus.61 Although the process and machinery are similar, they have very marked differences due, most likely, to an early diversification in the evolutionary process, proof of this is the fact that Arabidopsis thaliana has four Dicer-Like (DCL) proteins, 10 AGO and 3 functional RNA-dependent RNA polymerase (RDR) genes.57,62 Mutations in genes of the enzymatic biogenesis machinery, such as dcl1, hen1, hyl1, ago1, hst, are known to have dramatic effects on plant development.56 The biogenesis of miRNAs in plants, as in animals, begins co-transcriptionally63 with the recognition of the MIR gene by RNA Pol II, the only polymerase that to date is known to transcribe all MIR genes in plants.64 Once bound, RNA Pol II undergoes a phosphorylation-dephosphorylation process in the C-terminal domain of the large subunit. This process is mediated by the proteins CPLs (C-TERMINAL DOMAIN PHOSPHATASE-LIKE 1 AND 2) and CDKs (CYCLIN-DEPENDENT KINASES), which activate the transcription process and the transcript undergoes a self-folding process, thus forming the stem-loop known as pri-miRNA; the 5p end is methylguanylated and the 3p polyAdenylated.65–67 This nascent pri-miRNA is recognized by a type III Dicer-like endonuclease, the RNA-binding protein, HYPONASTIC LEAVES 1, the Zinc finger protein SERRATE and the RH27 (DEAD-BOX RNA HELICASE 27) protein, which together, they form a microprocessor that, in a first step, cuts the 5p and 3p ends close to the base of the pri-miRNA stem-loop, and in a second step, this same enzymatic microprocessor, cuts the pre-miRNA loop, thus yielding an RNA duplex.68,69 Next, this duplex is methylated at both 3' ends by the enzyme HUA ENHANCER 1 (s-adenosyl-L-methionine-dependent RNA methyltransferase, HEN1), promoting its stability and being the signal label for the enzyme HASTY (exp5 homolog of animals36) recognize the duplex and can export it to the cytoplasm, sponsored by the TREX-2 protein, which from the beginning of transcription joined Pol II;70 once in the cytoplasm, strand 5p is loaded into the RISC complex, which contains the ribonuclease AGO1 (strand 3p can be degraded or loaded into exosome-like microvesicles);71 the mature miRNA will guide to the RISC complex towards a target mRNA to which it will bind by nucleotide complementarity, thus inducing the AGO1 protein to cleave the mRNA70,72,73 (Figure 2B). The base complementarity between the 3' region of the mRNA and the 5' of the miRNA is the key point for the regulation or repression of the messenger that will be translated into protein;74–76 Generally in plants, complementarity is complete and the cleavage site by AGO1 occurs near the center of the miRNA complementary site (Figure 1B), specifically between residues 10 and 11 of the miRNA.34,77
There are other proposals for the biogenesis of miRNAs, in which the first stage of the biosynthetic pathway occurs within specialized processing bodies in the cell nucleus of plant cells, the so-called Dicing bodies (D-bodies),78–80 and once exiting the D-bodies, the miRNA duplex completes its maturation inside or outside the nucleus, including the release of the 5p strand of the duplex, to be exported to the cytoplasm in a single-stranded form with a degree of methylation.78 The participation of miRNAs in plants is a critical point for their proper development, for example, in tomato, miR171 participates in the branching of shoots, in the morphogenesis of the leaves and in male development, specifically in the pollen grain.81 Other miRNAs are involved in responses to abiotic factors, such as miR1320, Osa-miR156, Osa-miR319 and Osa-miR528, whose overexpression in rice confers greater resistance to cold. They are also involved in the development of organs or the flowering time of plants, such as miR172, whose overexpression in Arabidopsis accelerates vegetative development, also accelerating the flowering process.10 It is known that when caterpillars of the geometrid lepidopteran (Ectropis oblique) attack the Tea plant (Camellia sinensis), it responds by secreting volatile compounds that attract natural predators of the insect, and when these responses are activated, some miRNAs are differentially expressed, such as such as csn-miRn30, csn-miRn36 and csn-miRn40 that are expressed up, while csn-miRn68, csn-miRn109 and csn-miRn198 are expressed down, as an indirect response to the stress caused by geometrid.82 During the formation of adventitious roots in False Acacia (Robinia pseudoacacia L.), miRNAs control hormone signaling and starch and sucrose metabolism by regulating their target genes, some are highly expressed, as is the case of miR6171 and miR3267 that are increased during the first hours of root development, while others, such as miR477-3p, are down-expressed.83
miRNAs are involved in the production of flavonoids, as is the case of miR156 and miR858 from Brassicacoraphanus, where miR858 targets MYB transcription factor genes, and miR157 targets SPL (SQUAMOSA-promoter binding protein-like) genes, involved in the biosynthetic pathway of flavonoids, among which flavonoels, flavones, isoflavones and anthocyanins stand out.84,85 In poppy (Papaver somniferum), miRNAs can regulate the biosynthetic production of alkaloids; among the main miRNAs are pso-miR13, pso-miR2161 and pso-miR408.86 In the Indonesian cinnamon tree (Cinnamomum burmanni), the miRNA families miR396, miR5185, and miR9408 regulate the synthesis of mono and sesquiterpenes during leaf development. In particular, nobel_miR_377, belonging to the miR5158 family, reduces the expression of the enzyme 1-deoxy-D-xylulose-5-phosphate synthetase (DXS) involved in the 2-C-methyl-D-Erythritol-4-phosphate (MEP) pathway that, together with the mevalonate pathway, give rise to the biosynthesis of terpenes and terpenoids.87
Biogenesis and function of microRNAs in fungi
It is known that, in fungi, there are small non-coding RNA molecules similar to plant and animal miRNAs, miRNAs-like or milRNAs (miRNAs-like RNAs).88–90 These molecules were first described in Neurospora crassa in 2010 and later in Metharizium anisopliae, Sclerotia sclerotiorum, Penicillium marneffei, Trichoderma reesei, Antrodina cinnamomea, Fusarium oxysporum, Aspergillus flavus and Penicillium chrysogenum.91–93 Data analysis reveals the presence of Dicer and Argonaute proteins in N. crassa, A. nidulans, A. fumigatus and Paracoccidioide brasiliensis involved in the silencing mechanism mediated by small interfering RNAs (siRNAs), as well as by miRNAs, this indicates that the same machinery generates both species of RNAs.91 To date, there is no determined mechanism for the biogenesis of miRNAs-likes in fungi (Figure 2C), but it is known that the production of some such as milR-1 in N. crassa is dependent on Dicer and Argonaute qde- 2p (quelling deficient 2), while the biogenesis of other genes such as milR-2 is independent of Dicer. Unlike plants, the Argonaute protein is required for the production of pre-milRNAs and mature milRNAs in N. crassa.91 The existence of two DCL genes in P. marneffei is known; dcl-1 and dcl-2, as well as an Argonaute protein qde-2. In this fungus, both Dicer contain 4 domains that characterize Dicer family proteins; two RNase domains in the C-terminal region, a DEAD-box ATP-binding domain in the N-terminal domain, and an RNA helicase and double-stranded RNA-binding domain in the central region.92 Furthermore, the Argonaute protein possesses two domains that characterize the Argonaute family, namely, PAZ and Piwi, as well as the domain conserved in most Argonaute, DUF1785.91 Although all AGO family proteins in C. cinerea have been predicted to have at least one of the two PAZ or Piwi domains, the PAZ domain, which is a conserved domain in Argonaute proteins, could not be detected in C. cinerea, but it was observed that one of the DCLs in this microorganism contains the PAZ domain, which is present only in fruiting body-forming fungi, but not in other fungal DCLs, this suggests that the Dicer proteins diversified and duplicated at an early stage in the eukaryotic lineage, and the loss of the PAZ domain in DCL forced fungi to adopt different mechanisms for the production of milRNAs, resulting in milRNAs of different length sizes in fungi since the PAZ domain recognizes the 2nt of the overhang in the 3' end of the pre-miRNA in the biogenesis process,90,93 as occurred in thermal dimorphic fungi, where it is suggested that the dcl-2 gene may have co-evolved to fulfill DCL-like functions in plants and animals.92
There is evidence that some species of fungi that infect plant cells can send miRNAs to regulate host genes, for example, the fungus Botrytis cinerea, transfers small RNAs that bind to the host's AGO protein mRNA, thus suppressing host immunity genes.89 It is known that Metharrizium anisopliae contains the machinery of the RISC complex, including homologs of Dicer and Argonaute, this implies that miRNAs can be generated by the action of Dicer, but they were called milRNAs, since M. anisopliae lacks sequences homologous to Drosha and Pasha, miRNA biogenesis proteins in animals responsible for obtaining pre-miRNAs from pri-miRNAs.90 pre-milRNAs can be in the range of 68-208 nt, with an average of 126 nt.90 In Cryptococcus neoformans, the approximate size of the miR1 and miR2 precursors is ~70 nt.91 On the other hand, milRNAs may have different expression levels during different stages of development or in conidiogenesis. Some maintain their basal expression level, others are deregulated, and still others are upregulated, as is the case of man-milR-5, which expresses a high abundance during conidiogenesis of M. anisopliae, influencing the transcription factors for this process.90 milRNAs can be produced in fruiting body-forming fungi in routes similar to those of plants and animals, for example, cci-milR-12c controls hyphal growth by targeting the fungal pheromone, the hydrophobin of this stage, and the nucleotide metabolic process in basidiomycetes.93 It is also known that different Dicer and Argonaute proteins may be involved in the production of milRNAs in the different phases of dimorphic fungi, such as the case of dcl-2 involved in the production of milRNAs in the mycelial phase of P. marneffei.91 In the case of P. marneffei, the differential expression of PM-milR-M1 and PM-milR-M2 in the mycelial phase shows that it is dependent on dcl-2 but not on dcl-1 or qde-2, and the level of expression of dcl-1 is 25 times higher in the yeast than mycelial phase. The expression of dcl-2 and qde-2 was 7 and 2 times higher in mycelium than yeast, respectively.92 The approximate size of the pre-milRNAs of PM-milR-M1 and PM-milR-M2 is 70 and 91nt and the presence of both milRNA* and the 2 nt overhang at the 3' end in the milRNA/milRNA* duplex It is a strong indication that they are produced by Dicer-like as occurs in miRNAs. The size of M1-milR-M1 is 21 nt and it is suggested that DCL-2 is required for the production of PM-milR-M2, and that DCL-1 is not required for the production of milRNAs.92 It is known that, during a mycosis, the host activates the production of miRNAs in response to the pathogen, such as miR-212 and miR-132, which are expressed in response to A. fumigatus and C. albicans. During the germination of A. fumigatus conidia in lungs, different expression levels of both miRNAs and mRNAs can be observed in the host, as well as the repression of miR29a-3p, miR-30c-5p and their targets Clec7, SMAD2/3 and TGF. Clec7a emits a NON-inflammatory response, as it recognizes β-glucan from the hyphal cell wall, and SMAD family transcription factors are included in the TGF signaling pathway. By repressing these miRNAs involved in the regulation of these responses to A. fumigatus, host mRNAs involved in host inflammatory responses are allowed to be expressed.91 Likewise, it is hypothesized that P. marneffei may encode milRNAs involved in dimorphism and that they are differentially more expressed in mycelium than in yeast.92
Biotechnological applications of miRNAs
The field of biotechnological applications of miRNAs is increasingly promising, since these small molecules offer solutions to health problems, as well as nutritional and economic problems worldwide. In the case of animal miRNAs, one of the applications that is gaining more and more strength is their use both for the detection of cancers, as well as in the categorization of carcinomas, the fight against chronic and autoimmune diseases, and applications in the design of human nutritional charts. In some types of cancer, miRNAs can be encapsulated within exosomes, as in the case of retinoblastoma, and can be used as biomarkers for their detection.94,95 The use of miRNAs as cancer biomarkers96 is based on the detection of alterations in expression patterns, as is the case of miR-21, which is overexpressed in ovarian, lung, gastric, and breast cancer, colorectal, B cell lymphoma, and glioblastoma. And on the contrary, miRNAs that are deregulated are known as oncomiRs, such as miR-9, which reduces its levels in gastric, colorectal and hepatocellular cancer. Furthermore, some miRNAs are deregulated in one type of cancer and upregulated in other types, such as the case of this same miRNA, miR-9, which is deregulated in lung cancer, but is upregulated in breast cancer and neck cancer.97 Regarding the biotechnological applications of miRNAs in plants, these regularly focus on increasing crop yields, mainly focused on the availability and assimilation of nutrients,98 or resistance to pathogens.99,100 One of the most important crops globally is rice, in which miR171b has been overexpressed, resulting in plants that are more resistant to the fungus Magnaporthe oryzae, responsible for the disease known as blight, pyriculariosis or rice burn, a disease characterized by damaging the leaves of plants, causing a reduction in photosynthetic capacity, thus reducing production yields worldwide.101 Furthermore, plant miRNAs are being investigated for their potential use in dietary therapies, since, unlike other types of RNA, mature miRNAs have the ability to cross from one kingdom to another through diet, as is the case of rice miR168a, which was found in organs and the circulatory system in humans and mice.102 In health matters, mir2911 from the Japanese honeysuckle plant (Lonicera japonica) inhibits H5N1, H7N9 and H1N1 viral activity by decreasing the expression of the PB2 and NS1 protein encoded by H1N1.103,104 This same miRNA was able to inhibit the replication of the SARS-CoV-2 coronavirus, and accelerate the conversion of infected patients.103,105 With regard to fungal miRNAs, and in the same way as plants, these can be used biotechnologically to improve the yields of agricultural products by favoring beneficial plant-pathogen interactions, as in the case of the ectomycorrhizal fungus Pisolithus microcarpus that can regulate plant gene expression, using Pmic_miR-8 to attack and colonize the plant by repressing CC-NLR (CC nucleotide binding and leucine-rich repeat domain immune receptors) transcripts, one of the molecules responsible for recognizing and responding to pathogenic attacks.106
miRNAs are RNA molecules between 18 and 25 nucleotides in length that regulate different processes within living beings by binding to a specific mRNA, to which, in the case of animals, they bind and interrupt protein translation, and in the case of plants, the mRNA is cleaved, also avoiding translation into protein, and in the case of fungi, the mechanism of action has not yet been proposed, although due to its characteristics, it must be similar to either the mechanism of animals or to the plant mechanism. Similarly, in fungi, a route for the biogenesis of milRNAs has not been proposed; however, it is known that in some fungi, such as N. crassa, the Dicer and Argonaute proteins are required for their production, furthermore in M. anisopliae, the machinery of the RISC complex is available, although the milRNAs in this fungus lack sequences homologous to Drosha and Pasha, which suggests that the biogenesis of fungi may be more similar to that of plants than that of animals. With regard to biogenesis in plants and animals, the big difference is that, in plants, the entire process occurs within the nucleus, while, in animals, half of biogenesis occurs in the nucleus and the other half in the cytoplasm. Finally, it is worth highlighting the importance of the biotechnological applications of miRNAs in fungi, plants and animals, since they are being used to improve health conditions, increase the yields of agronomic products, and optimize the production of secondary metabolites of commercial interest in food grade.
To Instituto Politécnico Nacional and the CONACYT for financing the development of the article.
The authors declare that there are no conflicts of interest.

©2024 Gutiérrez, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.