International Journal of
eISSN: 2573-2889


Research Article Volume 7 Issue 1
1Ambo University, college of Veterinary Medicine, Ethiopia
2Haramaya University, College of Veterinary Medicine, Ethiopia
Correspondence: Adem Hussein, Ambo University, college of Veterinary Medicine, Ethiopia, Tel +251960987630
Received: March 03, 2024 | Published: March 29, 2024
Citation: Hussein A, Abraha B. Bacterial CRISPR/Cas9 system as discovery of promising solutions for all health problems and advancement in bioengineering. Int J Mol Biol Open Access. 2024;7(1):49-56. DOI: 10.15406/ijmboa.2024.07.00167
Bacterial Clustered Regularly Inter Spaced Palindromic Repeats (CRISPR) Associated proteins in association with a short guide RNA discovered as a bacterial adaptive immunity in the 20th century nowadays became a breakthrough gene editing tool in the arena of biomedical science and bioengineering. Historically, it was first identified as how bacteria maintain its genome integrity through a targeted endonuclease of any exogenous invading genetic elements either from plasmid or viruses by storing a memory of the first infection, expression of spacers and interference. Recognizing how it works, humans nowadays, are able to reprogram it using computer databases and genetic engineering knowledge and tools to cut and edit the genome at a targeted site for cancer therapy, viral therapy, and gene disruption for bioengineering purposes. They were also able to deactivate its endonuclease properties and making it only bind to the target site and act as a reporting signal in cooperation with other chemicals to indicate the presence of the genome so that it is being used as a diagnostic procedure for several diseases. However, the role of the system in repairing the broken DNA is unexplained or null. So in this detailed review, the historical discoveries, the mechanisms, processes, challenges and future research focus of the CRISPR/CaS9 how the system will be harnessed and un erroneously repairs the introduced break are discussed.
Keywords: bacteriophage, Cas9, CRISPR, CRISPRidCas9, double strand break, extra genomic element, Guide RNA, PAM
BP1, The tumor suppressor P53-binding protein 1; BRCA, breast cancer antigen; CAR, chimeric antigen receptor; Cas9, CRISPR associated protein9; Cascade, CRISPR-associated complex for antiviral defense; Cmr, Cas module RAMP (repeat-associated mysterious proteins); Cmr III-B, multiple subunit type III B CRISPR RNA-Cas protein; Cpf1, CRISPR from Prevotella and Francisella1; CRISPR, clustered regularly inter spaced palindrome; crRNA, CRISPR RNA; Csm III-A, multi-subunit Type III-A CRISPR-Cas protein; dCas9/sgRNA-SG I, deactivated Cas9/short guide RNA-Sybrr-Green I; DNA-PK, DNA-protein K; DNA-PKcs, DNA protein K catalytic subunit; DSB, double strand break; EGE, extra genic element; gRNA, guide RNA; HDR, homology directed repair; IAP, alkaline phosphatase isozyme; MRE 11, meiotic recombination 11; NHEJ, non-homologous end joining; PAM, proto-spacer adjacent Motif; PD, programmed cell death; RAD, recombinase A; REP, repetitive extragenic palindrome; RPA, replication protein A; RT, reverse transcriptase; sgRNA, short guide RNA; SSB, single strand break; tracrRNA, Trans-activating CRISPR RNA; XLF, XRCC4-like factor; XRCC 4, X-ray repair cross complementing protein 4; YOYO-1, (Oxazole Yellow)
Bacteriophages are the most abundant nature of life (viruses) known to infect bacteria. They out-number bacteria and are very deadly for them. Bacteria do acquire extra genic mobile elements (EGE) ((free DNA, plasmid) as an evolutionary process or phage (when they are infected)) through transformation, conjugation, and transfection. But infection by bacteriophages or horizontal genomic acquisition by bacteria is possible only in non- CRISPR/Cas systems containing bacteria.1 So, since the discovery bacterial adaptive immune system against any invading EGE by,2 human perception regarding bacterial EGE acquisition from the environment through whatever mechanism and development of antibiotic resistance is totally changed. Sometimes, nature provides a full package of living requirements. But it is up to us to discover it and use it to solve our entire genetic-related problem, diagnostic, production, and performance trait improvement, or all about specific and efficient gene editing requirements. This is the breakthrough of the CRISPR/Cas9 gene editing invention in bacteria.
CRISPR/Cas9 system is an acronym standing for Clustered Regularly Inter Spaced Palindromic Repeats/ CRISPR Associated protein9.3 It is the only adoptive immunity described yet in prokaryotes against any invading EGE (bacteriophage, plasmid, or transposons) in the protection of their genomic integrity. In this system, short guide RNAs (sgRNA) called CRISPR RNA (crRNAs) transcript from parts of previously invaded nucleic acids are employed for sequence-specific cleaving of any integrated EGE by Cas protein. CRISPR-Cas system in bacteria and archaea has an operonic genomic locus comprising three separate sequences; these are an AT-rich leader sequence flanked by a set of Cas genes encoding the Cas proteins, with palindromic repeats interspaced by unique spacers of bacteriophage, plasmid, or any EGE origin.4 Today, beyond bacterial adaptive immunity, CRISPR/Cas system is being employed as a fast, efficient, and specific gene editing tool to solve many problems. For instance, a study undertaken by Rothemejer et al.5 indicated that CRISPR/Cas9 edited and single chain variable fragment (scFv) neutralization antibody CAR cassette integrated T cells at the CCR5 locus showed HIV resistance, expansion, and specific lysis of HIV-infected cells in- vitro and in a humanized mice. So, understanding how the CRISPR/Cas9 system functions in bacteria and be reprogrammed to use in animals genome editing for several purposes is an uplift. Generally, this provides brief insight on CRISPR/Cas9 regarding its historical development, how it works, and how it could be reprogrammed to use in therapeutic and diagnostic applications and its future perspectives.
The CRISPR/CAS systems
History
Since 1984 and before, the repetitive gene sequence called REP is known among the scientific community. Stern et al.6 have explained this more in detail about their abundance and homology, over a stretch of about 35bp. According to their work REP sequences are present in 26% (16/61) of all the E. coli and S. typhimurium operons they have examined. In 1987 unusual structure in the sequence E. coli was also discovered by Ishino and his colleagues during the sequencing of E. coli DNA fragments containing the IAP gene. In this study, they got five highly conserved sequences of 29bp that were arranged as direct repeats with 32 nucleotides as spacers. All of these sequences are known to be found in all prokaryotes and archaea at non-translated regions, either between two genes that are co-transcribed as part of a single operon or, alternatively within the 3’ un-translated region at the end of an operon.6,7 While Stern et al.6 wrongly claim a possible role of REP sequences as a regulator of gene expression, Ishino et al.7 said the biological significance of REP sequences is not known. Hermans et al.8 discovered these direct repeats in Mycobacterium bovis and suggested it as one of typing roles in Mycobacterium.
The greatest move in the discovery of CRISPR/Cas9 was the works of a Spanish microbiologist Francisco Mojica. Since 1993, Mojica was working with different halophilic archaeon and prokaryotes genome sequencing and identified a peculiar repetitive DNA sequence in their genome which later he called Short Regularly Spaced Repeats (SRSRs). 9–11 In collaboration with Mojica, Jansen et al.3 called these sequences the CRISPRs for the first time, after they discovered the Cas gene and its functional relationship with CRISPR (helicase and nuclease) at the loci. According to his discovery, an organism may contain two or more CRISPR loci containing 300-500 nucleotide leader sequences and the direct repeats sequences that were conserved within a species, but dissimilar between species. Jansen in his 2002 research reported four different Cas (named from 1-4) protein loci working together as a cascade for its function. These novel repetitive DNA sequences are absent from eukaryotes or viruses.3
Later Bolotin et al,12 Mojica et al,13 Pourcel et al,14 and Deveau et al,15 showed that these repeats have 21-37bp and are spaced by a non-repetitive sequence of 29-38bp of phage origin. However, till Barrangou’s et al,2 experimental demonstration of CRISPR-Cas systems as acquired immunity of bacteria and archaea against bacteriophages, the functionality of CRISPR locus was unclear except for Mojica’s hypothesis stating the CRISPR/Cas locus functionality might be controlling of both mobility and maintenance of EGE that could be from phage. But regarding its possibility, the explanation was credited to Brouns et al,16 when they discovered that prokaryotes acquire virus resistance by integrating short fragments of viral nucleic acid into CRISPR locus then transcribing and maturing it through cleaving and using as short guide RNAs (crRNA) for viral derived gene surveillance of Cas nuclease domain. In the same year, Marraffini and Sontheimer,1 showed that CRISPR loci can interfere with multiple routes of horizontal gene acquisition of bacteria which can limit the spread of antibiotic resistance in pathogenic bacteria.
As a completion of the discovery of the CRISPR/Cas gene regarding what it is and how it works, Deveau et al,15 identified a phage phenotype called proto-spacer, which is important for the Cas system to recognize the spacers. Later this sequence is called proto- spacer adjacent motif (PAM). Further Deltcheva et al,17 discovered tra-crRNA a 24- nucleotide base complementary to the crRNA precursor, that directs the maturation of crRNAs by the activities of the highly conserved endogenous RNase III and the CRISPR-associated protein 9 (Cas9); all these components are inactive alone. The breakthrough started here when Sapranauskas et al,18 showed the transferability and proper functionality of the system in genetically distant organisms. Finally, Jinek et al,19 showed that the Cas9 endonuclease can be programmed with synthetic guide RNA engineered as a single chimera transcript to target and cleave any dsDNA sequence of interest. Later in 2020 Jennifer Doudna and Emmanuelle Charpentier got a Nobel Prize in the area of CRISPR/Cas9 genome editing and shared with Mojica Figure 1.
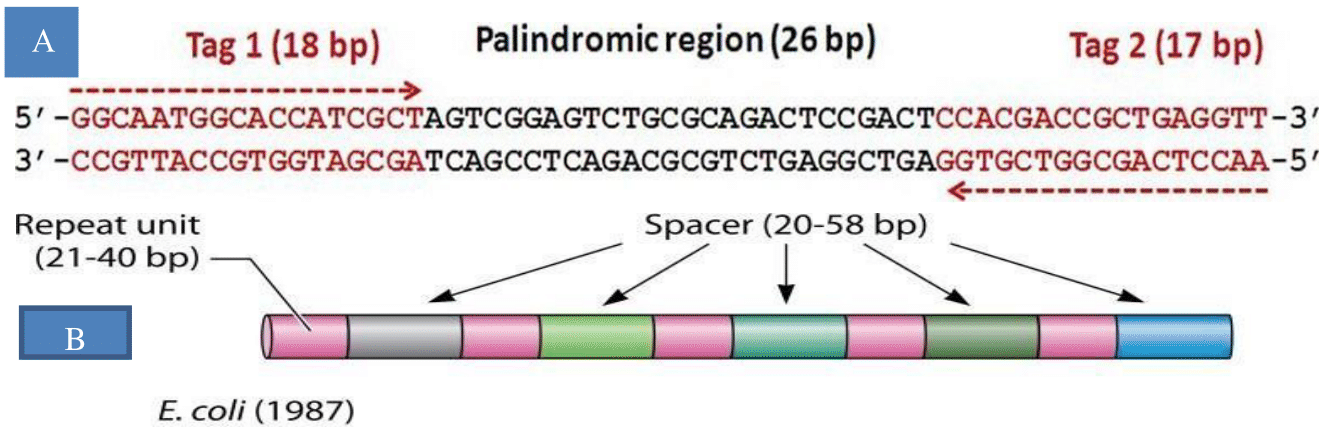
Figure 1 (A) A 26-bp palindromic sequence flanking two tags originated from the E. coli genome (B) a computer model of its arrangement.
Types and mechanisms of CRISPR/Cas system
To date, there are two classes of CRISPR/Cas system which are further divided into six types and in turn subdivided into 33 subtypes.20 The basic difference between the classes is the number of effector modules (expression and maturation of the guide molecule and cleaving of the target). Class 1 (type I, III, and IV) involve a cascade of several proteins for these functions. Whereas, class 2 (type II, V, and VI) involve single multi-domain proteins Cas9, Cas12, and Cas13 respectively with matured short guide RNA Table 1.21
|
Basis of classification |
Type I |
Type II |
Type III |
Type V |
Type VI |
|
Cleaving protein |
Cas3 |
Cas9 |
Cas10 (Csm and Cmr subunits |
Cas12 (a(Cpf1), b, c, d, and e) |
Cas13 (a, b, c) |
|
PAM sequence |
Any 3-nts |
5’------ NGG-3’ |
Without PAM |
5′-TTTN-3’ |
Without PAM |
|
RNA guide |
crRNA and cascade of Cas systems |
crRNA: tracrRNA |
crRNA and cascade of Cas systems |
crRNA and tracrRNA for 12 b |
crRNA/PFS |
|
Target molecule |
DNA |
DNA |
DNA/RNA |
DNA/RNA |
ssRNA |
|
Product |
SSB forming |
blunt-ended DSB |
SSB |
5 to 7 nt overhang DSB |
ssRNA |
Table 1 The basic difference between different types of CRISPR/Cas systems
Mechanism of CRISPR /Cas9 workflow
The whole CRISPR/Cas system works in three sequential and independent steps known as Adaptation or Spacer acquisition, expression or maturation (pre-crRNA processing), and Interference or effector function step. Based on these functions, the CRISPR-Cas system genomic loci were assembled in three consecutive iterations. These are the adaptation module (cas1, cas2, and/or cas4 and/or RT), the expression module (cas6 for all class 1 Cas systems, RNase III for class 2 type II, and Cas 12 and Cas 13 for class 2 type IV and VI respectively). And finally target guiding and cleaving module are cas9, cas12, and cas13 for CRISPR-Cas Class 2 type II, IV, and VI and cas3, cas5, cas7, cas8, cas10, cas11, and others are assumed for Class 1 even though not fully understood Figure 2.20
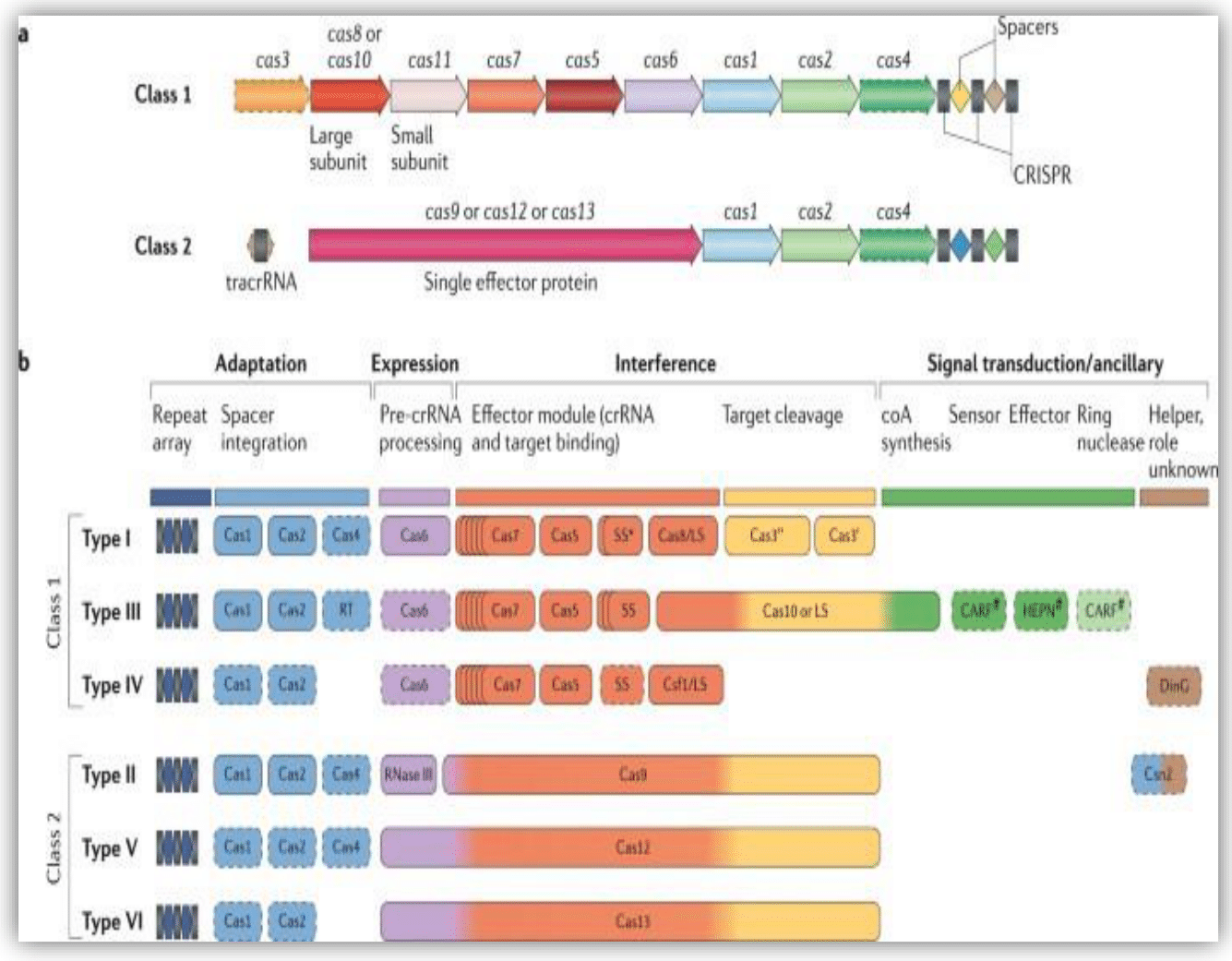
Figure 2 The different classes and types of CRISPR/Cas system with their gene loci responsible for different effector functions of the Cas system.
As indicated in part “a” the generic organizations of class 1 and class 2 CRISPR–Cas loci, the bacterial CRISPR/Cas genome organization is an operon containing Cas genes coding for different Cas proteins found nearby a repetitive array of palindrome interspaced with different sequences from any EGE that historically infected the bacteria. Part “b” indicates the functional organizations of CRISPR–Cas systems in different classifications. All types of Cas systems contain repetitive arrays, spacer adaptation modules, the pre-crRNA processing, and maturing module, and finally the interference module. The main difference between these Cas system classes and types is the protein responsible for such modules as indicated by different colors. And still, some may contain additional features that function not understood yet as indicated by the pound symbol (#).
Spacer acquisition/adaptation
Assumptionally, all CRISPR/Cas system-positive bacteria and archaea contain naïve CRISPR loci; i.e., their CRISPR locus did not incorporate any spacers at the first time; rather it only contains the Cas gene with palindromic repeats.4 Through evolution from this time, the bacteria’s Cas system adaptation module (cas1, cas2, and/or cas4 and/or RT) scans through the genome of any invading EGE (RNA or DNA) in search of parts that accurately represents the invading genome containing the PAM sequence the NGG for Cas 9.22–24 Once it gets the correct sequence, the module cut and integrates it into its CRISPR locus. Later on, the bacteria do not need to integrate the spacers from the same origin for all the coming generations. Simply it will recognize the invading genome through its crRNA transcription.
Expression/maturation
This step in the workflow of the CRISPR/Cas system is the step in which the short guide RNA (sgRNA or crRNA) containing memorized sequences of invaders is fully functionally prepared from its long precursor crRNA (pre-crRNA) that is generated from the transcription of the CRISPR locus. This is through pre-RNA processing module endonuclease activity of Cas12 and Cas13 in type V and type VI respectively, and Cas6 in type III, IV and type I Cas systems other than type I-C. Specifically in the type I-C Cas system, Cas5d processes the pre- crRNA; producing 2 and 11 nucleotides (nt) overhangs at the 5’ and 3’ ends, respectively in a metal-independent manner capable of guiding the cascade for further interference.25,26 In the type II system, host RNase III process up on Cas9-RNA duplex (tra-crRNA anti-repeat sequence-repeat sequence pre-crRNA) recognition.4
Interference
The last steps in CRISPR/Cas system bacterial adaptive immunity against any invading EGE is the specific recognition of the PAM sequence and “seed (7-nts directly next to PAM) in a spacer” through Cas Protein-tracrRNA-crRNA complex and introduction of DSB at specified place upstream of the PAM.4 Mutation in the PAM and seed sequence will invalidate the CRISPR-based bacterial immune system.27 Even if it is not well established for all types of Cas systems, many researchers Semenova et al,27 Shah et al,28 and Westra et al,29 discovered the need for proper match of the PAM (upstream for type I and V or downstream type III to the protospacer) and seed sequence for the function of this system and to avoid autoimmunity. In the type II CRISPR Cas system, an additional requirement is the host-derived 5’ tag (8 nt from repeat sequence) that provides for discrimination between self and non-self.30,31 The PAM sequences are highly varied among different types of CRISPR systems and even within the same system among different spacers.32
The recognition of the very essential module of the invaders: the PAM (for type I, II, III, V), PFS (for type VI), and 5’ Tag (additionally for type II) and cleavage of the invader is undergone by the CasCADe of Cas proteins that have a function of guiding crRNA to the target, R- loop stabilization, PAM recognition, and cleavage.33 The functions assigned to these Cas systems are different in different Cas systems. In opposed to this, the type II Cas system employs a single bi-lobed Cas 9 protein as a recognition of the PAM and cleavage when but only cooperated with guide crRNA and recruiting tracrRNA (dual RNA chimera).19 Upon binding to the PAM sequence and subsequent guide RNA–spacer DNA duplex formation, the Cas9 enzyme is activated and introduces blunt DSB at a specific site 3 bp downstream through the spacers by its RuvC active site (within complementary strand) and HNH active site (within non-complementary strand) Figure 3.34
When the invader (plasmid or virus) enters bacteria, it directs a nuclease called Cas2 to scan and snip a short sequence of the viral genome (spacer) and insert it between two repeats in its CRISPR locus. When this invader type comes again, the bacteria transcribe its spacer to generate crRNA, which will be matured by tracrRNA. Both types of RNA are associated with Cas9 and will be directed to the invader genome to cleave it (using Cas9) after PAM recognition and direction from crRNA.19
Reprogramming for use in CRISPR negative organism
Guide tracrRNA-crRNA chimera plasmid generation
To use guide RNA (gRNA) programmed Cas9 endonuclease as a tool for problematic gene locus editing in health or genetic engineering for the high-performance trait, both Cas9 vector which could be plasmid, and engineered gRNA with its scaffold must reach a fully functioning nucleus. So, these components can be introduced to cells, either as a DNA, an in-vitro mRNA transcript or a protein form for Cas9 and a DNA plasmid, or as an in-vitro mRNA transcript for guide RNA.35 The programming of this breakthrough but infant, bacterial genome editing system for use in eukaryotic cells is begun by the synthesizing a plasmid or vector carrying computer database programmed-laboratory made tracrRNA-crRNA guide oligo sequences and Cas9 gene with their promoter. The oligonucleotide must perfectly mimic the in vivo phenomenon through the usage of intended gene-specific promoter and guide sequences, starting codon and stop codon and Cas9-specific tracrRNA. For this, according to Yang et al,36 a 22-bp region within 50-bp in the target genome, which may be on either of the strands and must overlap the target sequence is identified in the form of 5’- A/G and sometimes C -N19-NGG-3’ using sequence analysis software BLAST. In this design, while the 3’ NGG is the protospacer adjacent motif (PAM) that is critical for Cas9 recognition of the target.4 the A/G and sometimes C base at -1 to +1 position is necessary for the U6 promoter to initiate transcription of the gRNA plasmid.37 (Ma et al., 2014). With minimal alternate targets on the genome, at least the 13 bp before NGG protospacer adjacent motif (S13NGG) on the 3’ must accurately match the reference sequence.19
Finally, a design incorporating the promoter of gRNA, specific gRNA, gRNA scaffold, and stop codon is made in the laboratory and inserted into the Cas9 plasmid containing selectable markers (according to Figure 4 below). The plasmid is screened for an insert and those accurately expressing the desired insert are used in the transfection of initiated cell line or embryonic stem cells in an appropriate condition for infusion (for example CAR-T cell therapy), germinal, or somatic cell-specific genome site edition.
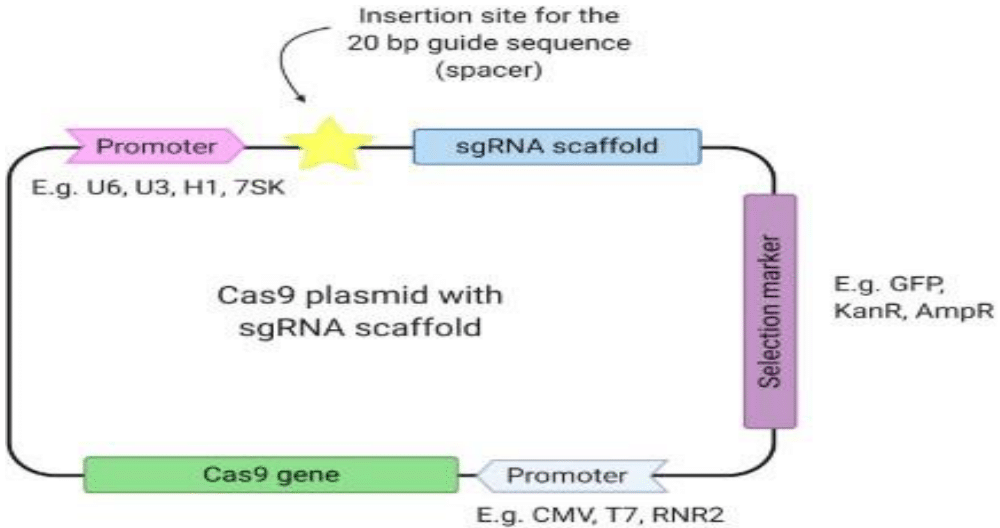
Figure 4 Cas9 expression vector plasmid containing promoters, insertion site for 20 bp guide RNA (spacer), a sgRNA backbone, the Cas9 sequence, and a potential selection marker gene.
DNA double-strand break repair mechanisms
Many researchers Jinek et al,19 Jinek et al,38 and Mali et al,39 have shown the capability of RNA-programmed Cas9 system engineering and transfect-ability to introduce DSB in the genome of many cultured eukaryotic cells including that of humans at specific sites intended to be edited. However, none of them explained the system’s role in repairing the broken DNA. Under so many circumstances DSB in the cellular genome happens at an estimated rate of 10 DSBs per day per cell;40 even if it rarely resulted in a harmful effect like cancer and death of the cells. As a reason, scientists discovered how the broken genome is repaired and integrity is maintained and passed to the next generations without any external interventions.41 This discovery is the basis for CRISPR usage, as it only introduces the DSB within the genome at a specific site intended to be edited and it is up to the cell to repair the double-strand break.42 In all life being either eukaryotic or prokaryotes or archaea, cellular DNA DSBs that are introduced in many circumstances are rejoined through homology-directed repair (HDR), and/or non-homologous end joining (NHEJ) pathways.
The NHEJ pathway is an error-prone and default DNA repair pathway in mammalian cells; probably because of insufficient homology. Whereas, HDR is efficient and error-free which is dominant in bacteria.42–44 Homology-directed repair is a cell cycle (S/G2 phase) and replication-dependent double-strand break repair mechanism that uses either undamaged sister chromatids or any other homologous chromosome from vectors or viruses.44,45 The complexity of the damage and the presence of homologous chromosomes are the factors that switch NHEJ to homologous recombination.45 Proteins involved in HDR (Homologous recombination) include RAD51, RAD52, RAD54, RAD55, RAD57, RAD50, and MRE11.44 It is initiated by MRE11-dependent resection of double-stranded DNA ends and extension of 3’ single-stranded DNA overhang of the resected ends by positioned 53BP1 via a BRCA1- dependent process. Then the 3’ends filament are bounded with replication protein A (RPA) which would be replaced by RAD51 that promotes the invasion of the undamaged homologous strand, and D-loop formation generating a heteroduplex molecule that results in the synthesis of the resected part and ligation of the double-strand breaks.45
On the other hand, NHEJ (non-homologous end joining) is active throughout the cell cycle, faster than HDR, and suppresses the HDR process. It is initiated by the binding of Ku70-Ku80 protein at broken sites which serves as a scaffold to recruit other NHEJ-related factors like DNA-PKcs and protect the DNA damage sites from end resection by exonucleases. The DNA- PKcs recruitment stimulates the kinase activity of DNA-PK and orchestrates NHEJ through phosphorylation of Artemis, X-ray repair cross-complementing protein 4 (XRCC4), DNA ligase IV (Lig IV), and XRCC4-like factor (XLF) which promotes the synapsis of DNA ends and facilitates the recruitment of end-processing and ligation enzymes. By these processes, where Aligned and compatible DNA termini can be directly ligated by the NHEJ factors, incompatible DNA termini are prepared through DNA blunt end formation by Artemis, cooperating with polymerase lambda (λ) and mu (µ) that randomly add and remove DNA bases resulted in small indels relative to the original genomic template. Finally, the XRCC4- DNA ligase IV-XLF complex performs the ligation across gaps separately and independently of each other.46
Genes made up of a pool of DNA on a chromosome are known to be a code of life through its central dogma of life.47 Nowadays, over 3000 genes have been associated with disease-causing mutations.48 Approximately 8.5 million people died of cancer globally each year due to its high lethality and absence of effective therapy.49 Several genes in the host and pathogen have also been shown to code for receptors and ligands contributing to the virulence of different pathogens.50 At the same time, these genes code for some desirable trait that needs to be enhanced. Under this heading, we discuss how CRISPR/Cas9 gene editing tool act as cancer and viral therapeutic and immune, enhancement of production traits in food-producing animals, and diagnostic tools for diseases.
Therapeutics
Viral Therapy
HIV/AIDS
The clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated nuclease9 (Cas9) system is shown potentially eliminate or disrupt HIV-integrated genomes in HIV-infected cells from multiple HIV reservoirs, which could result in the complete cure of HIV/AIDS.51 Furthermore, as a receptor editing function, media all over the world reported the birth of an HIV-immune twin girl with CRISPR-Cas9-based genome editing, by disabling the CCR5 gene that enables HIV infection by He Jiankui in November 2018. Finally, his work was rejected due to research ethics disobedience. This work was driven by the work of Niu et al,52 indicated precise and multiplex one-cell-stage cynomolgus monkey embryos by co-injection of Cas9 mRNA and multiple sgRNAs. Additionally, a study undertaken by Rothemejer et al,5 indicated that CRISPR/Cas9 edited and neutralization antibody CAR cassette integrated T cells at the CCR5 locus showed expansion and specific lysis of HIV-infected cells in-vitro and in a humanized mouse.
Cancer Immunotherapy
B-cell Non-Hodgkin Lymphoma (B-NHL)
So many researchers have shown that chimeric antigen receptor (CAR)-T cell therapy has great promise in treating hematological malignancies.53,54 CRISPR-Cas9 engineered and clonally expanded laboratory CAR-T cells have been shown high rate (87.5%) of complete remission and durable responses without serious adverse events in the preclinical study by inserting an anti-CD19 CAR cassette into cytotoxic T cells through the integration of PD1 receptors in refractory aggressive B cell non-Hodgkin lymphoma (ClinicalTrials.gov, NCT04213469 ).55
Lung Cancer
Lu and co-workers performed a clinical trial-based experiment (NCT02793856) to investigate the feasibility and safety of CRISPR/Cas9 edited T-cell by co-transfection using electroporation of Cas9 and single guide RNA plasmids in patients with advanced non-small- cell lung cancer by targeting the PD-1 gene. In this first-in-human phase I clinical trial the median overall survival rate of 42.6 weeks (95% CI: 10.3–74.9 weeks with a short life span of edited T cells (7.7 weeks (95% CI: 6.9 to 8.5 weeks)) suggesting limited off-target effects indicate feasibility and safety of the system. From a total of 22 enrolled patients; 17 had sufficient edited T cells for infusion, 12 were able to receive treatment and edited T cells were detectable in peripheral blood after infusion. The off-target effect in 18 patients accessed by next-generation sequencing was 0.05% (95 CI: 0–0.25%).56
Gene Regulation
Deactivated CRISPR-Cas9 (CRISPRa and CRISPRi)
Deactivated (d) Cas9 is Cas9 with a nonfunctional endonuclease lobe. It has been used to up- regulate or down-regulate gene transcription when harnessed with a transcription activator that recruits RNA polymerase and a transcription suppressor which inhibits RNA polymerase binding respectively.56,57 This system can selectively regulate the expression of a target gene guided by sgRNA. In bacteria, CRISPRi is a more preferred transcriptional regulation tool for gene knockdown than any interference.57 According to Myrbråten et al,57 the CRISPRi system is ideal for quickly screening both essential and nonessential phenotypes genes.
Trait Improvement
Being a flexible approach for genome engineering of any genetic loci, the CRISPR/Cas9 system provides resilience against diseases, improvement of reproductive traits, and animal production by simultaneously targeting multiple genes that are responsible for economically significant traits.58 In a study by Wang et al,58 disruption of the MSTN gene in sheep at one-cell-stage embryos with Cas9 mRNA resulted in the desired muscle hypertrophy that is characterized by enlarged myofibers which accounts for meat improvement. The impact of CRISPR technology could potentially lead to the efficient improvement and sustainability of poultry products, which will help address challenges associated with universal food security. Birds raised for meat and egg production using the CRISPR technology could have an immense impact on the advancement of poultry-related traits such as feed conversion, digestibility, increased egg production, growth, and overall improved performance of birds.
Diagnostic
As indicated by Müller et al,59 combined CRISPR-Cas9 with optical DNA mapping tool to identify bacterial and any specific genes including antibiotic resistance genes. As of their study, a gRNA-Cas9 complex binds and cleaves gRNA-specific nucleic acid sequence of plasmids or genome containing resistance genes or genes of interest, and a fluorescent dye YOYO-1 (Oxazole Yellow) and netropsin independently bind DNA based on AT-rich region selectively, resulting in an emission intensity unique to each DNA segment, like a bar code. As a diagnostic system modified CRISPR/Cas9 (dCas9/sgRNA-SG I) based detection approach has proved to be easy, fast, sensitive, and cost-efficient. In this approach modified dCRISPR/Cas9 DNA fluorescent in situ hybridization (FISH) detects specific pathogens including methicillin-resistant Staphylococcus aureus (MRSA). The method utilizes a deactivated (d) Cas9 system where a sgRNA-dCas9 complex coupled with an SYBR-Green I fluorescent probe recognizes the specific gene of pathogens. While the complex recognizes the target DNA sequence, dCas9 does not induce DNA cleavage but remains in place making it suitable for detection by FISH.60
Multi-drug resistance inhibition
Other good news from the discovery of this sophisticated and wholesome bacterial adaptive immune system, the CRISPR/Cas9 is that it changed our perception of how bacteria acquire any EGE including plasmid, and become antimicrobial resistant. For this Barrangou and his colleagues in their experiment proved that CRISPR/Cas system containing bacteria are no longer become antimicrobial resistant by any horizontal gene transfer, as this system prevents and reverses any invading mobile genetic elements.2
In conclusion, bacterial clustered regularly inter-spaced palindromic repeats/ CRISPR Associated protein9/Cas9 system from its early discovery as a bacterial acquired immune system to modern targeted genome editing tools has passed through several assumptions and experimental studies. Being reprogrammable through exact mimicking of natural phenomenon by synthesizing DNA sequence containing a specific promoter, specific guide sequence of interest, PAM sequence with a stop codon, and scaffold having the ability to form a loop, CRISPR/Cas9 nowadays has been proved to solve many health problems and used in many bioengineering and diagnostic applications with some promising efficacy. These numerous applications of this system are due to its ability to introduce targeted DNA genome double-strand breaks guided by computer database-generated guide sequences. Yeah! As of all the articles for this review, CRISPR/Cas9 can introduce DNA double-strand breaks at specific sites with few off-target effects. However, the repairing of the broken DNA is solely given to the cells. Therefore, Further research on how to harness this system with homologous oligonucleotides and different DNA repairing enzymes to assist cellular DNA repair mechanism (favor HDR mechanism), and evaluating its clinical trials for its full adoption and applicability must be undertaken.
None.
The authors declare that there are no conflicts of interest.

©2024 Hussein, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.