International Journal of
eISSN: 2573-2889


Short Communication Volume 2 Issue 3
1Institute of Nutrition and Functional Foods (INAF), Laval University, Canada
2School of Nutrition, Laval University, Canada
3Quebec Heart and Lung Institute Research Center, Canada
4Department of Medicine, Laval University, Canada
5Department of Surgery, Laval University, Canada
Correspondence: Marie-Claude Vohl, Institute of Nutrition and Functional Foods (INAF) Laval University 2440 Hochelaga Blvd Quebec, QC Canada, G1V 0A6, Tel +1 (418) 656-2131 (4676), Fax +1 (418) 656-5877
Received: February 16, 2017 | Published: April 21, 2017
Citation: de Toro-Martín J, Guénard F, Tchernof A, et al. Bariatric surgery induces hypomethylation of genes related to type 2 Diabetes and insulin resistance. Int J Mol Biol Open Access. 2017;2(3):89-92. DOI: 10.15406/ijmboa.2017.02.00020
Biliopancreatic diversion with duodenal switch (BPD-DS) is a surgical intervention known to induce substantial weight loss and significant long-lasting metabolic improvements including a decrease in insulin resistance (IR) and resolution of type 2diabetes(T2D). The specific mechanisms by which metabolic improvements occur after BPD-DS are still not fully elucidated and the impact of BPD-DS on gene methylation profiles has not been studied. To gain understanding of epigenetic factors that may predispose to metabolic improvements after weight loss surgery, we characterized the methylation signature of genes associated to T2D and IR after BPD-DS. Most of the genes involved in T2D and IR pathways exhibited significant differences in methylation levels after BPD-DS compared to a pre-surgery control group. The majority of these loci were significantly hypomethylated, suggesting an effect of bariatric surgery on the epigenetic signature of genes encoding proteins involved in glucose homeostasis.
Keywords: CPG sites methylation, bariatric surgery, epigenetics, type 2 diabetes, insulin resistance
BPD-DS, biliopancreatic diversion with duodenal switch; IR, insulin resistance; T2D, type 2 diabetes; VAT, visceral adipose tissue; CpG, cytosine-phosphate-guanite dinucleotide; BL, blood leukocytes; KEGG, kyoto encyclopedia of genes and genomes
Biliopancreatic diversion with duodenal switch (BPD-DS) is a bariatric surgery inducing substantial weight loss and long-lasting metabolic improvements including, among others, improved insulin sensitivity and resolution of type 2 diabetes (T2D).1The specific mechanisms by which metabolic improvements take place after BPD-DS are still not fully elucidated. Among others, epigenetics has emerged as a novel mechanism potentially explaining part of the variability in the metabolic processes involved in glucose homeostasis.2
Knowledge of epigenetic factors determining the development of metabolic diseases remains largely incomplete. Previous studies have investigated the role of these factors in the severity of obesity-associated metabolic disturbances both at the whole-genome and candidate-gene levels. Previously, we reported that lower global methylation in visceral adipose tissue (VAT) may play a critical role in the development of obesity-associated metabolic alterations.3 We also revealed that the epigenetic signature of VAT differed between obese men discordant for metabolic disturbances,4 specifically within genes having a role in molecular pathways related to cell structure and cycle regulation, as well as in inflammation and immunity. We subsequently focused on two of the differentially methylated genes involved align those pathways, namely ARPC35 and TOMM20,6 revealing a role of both genes in lipid management among individuals with severe obesity, potentially driven by an epigenetic-mediated mechanism. Concretely, this might occur through a differential methylation at cytosine-phosphate-guanine (CpG) sites located within gene promoter regions. We found that methylation of these loci potentially altered plasma triglyceride and cholesterol levels.
Studies carried out in subcutaneous adipose tissue (SAT) gave rise to similar findings to the aforementioned works in terms of affected metabolic pathways. A recent study performed in twins discordant for T2D and in an independent cohort of unrelated patients, found both epigenetic and transcriptional changes in key genes involved in lipid (PPARG) and also in carbohydrate metabolism (IRS1).7 Likewise, fat cells from SAT of women with obesity also showed significant changes at epigenetic and gene expression levels mainly in adipogenesis, lipolysis and insulin signaling pathways.8 Significant DNA methylation differences have also been found in other tissues than VAT or SAT. Davegårdh et al.9 identified epigenetic and transcriptional changes in skeletal muscle between patients with versus without obesity. These changes were mainly observed in myogenic transcription factors, but also in other genes such as IL-32, which was further revealed as a novel target regulating insulin sensitivity. With a more precise focus on T2D, Dayeh et al.10 showed that the epigenetic signature of pancreatic islets of T2D-patients was significantly different to that of islets from healthy donors, mainly at genes involved in insulin secretion and glucose homeostasis.10,11 Altogether, these findings highlight the potential role of epigenetics in T2D pathogenesis and reveal that epigenetic changes taken place in obesity and T2D share common metabolic pathways and have a wide tissue distribution.
In this sense, we further broadened our research of VAT to more accessible blood leukocytes (BL). We hypothesized that some of the epigenetic determinants of BL associated with obesity-related complications may serve as surrogate markers of VAT.12 Results of that study suggested that BL epigenetic signature may adequately reflect VAT methylation levels for genes associated with the development of obesity-related metabolic disturbances, specifically for genes encoding proteins involved in inflammation, and the metabolism of lipids and glucose.
Whether epigenetic profiling could identify predictors for the heterogeneous response to weight loss surgery also represents a new, barely studied paradigm. We have previously shown that epigenetic changes in BL could be involved in the metabolic profile improvement after weight loss surgery. Specifically, we revealed that offspring born after maternal BPD-DS surgery showed differentially methylated genes predominantly involved in glucoregulatory, immune, inflammatory and vascular diseases13,14 Moreover, methylation levels, VAT mRNA abundance and markers of IR were significantly associated with metabolic improvements in offspring born after bariatric surgery.
More recently, to gain understanding of factors predisposing to metabolic improvements after weight loss surgery, we investigated the role of epigenetics in the resolution of T2D and related metabolic disturbances after weight loss surgery. Specifically, analysis of CpG sites methylation levels in women before and after BPD-DS revealed once again that diabetic and inflammatory/immune functions were among the most overrepresented in the list of genes with the largest methylation differences15 Herein, our aim was to deepen our understanding of these epigenetic determinants in the metabolic improvements observed after BPD-DS by identifying the methylation signature of genes encoding proteins associated with T2D and IR.
Briefly, whole-genome methylation levels at CpG sites were measured in blood DNA (Infinium Human Methylation 450 Bead Chips) of 20 women post-bariatric surgery (BPD-DS group) with a mean follow-up of 12years and in 20 pre-surgical women with severe obesity (control group). Participating women were matched for age, pre-surgery body mass index and metabolic parameters. A pathway-based differential methylation analysis was performed between the two groups for a set of genes encoding proteins involved in T2D and IR (T2D-IR genes), according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (IDs: 04930 and 04931). Differential methylation analysis was performed for 3,984 CpG sites, corresponding to 133 T2D-IR unique genes.16
Anthropometric and metabolic parameters of study participants are summarized in Table 1. A total of 811 CpG sites belonging to 119 T2D-IRgenes (89.5% of genes) remained significantly different after Bonferroni correction (P<1.43x10-7) (Figure 1a). Most differentially methylated CpG sites (60.8%) showed differences greater than 10%, and 97.2% of these sites were found to be significantly hypomethylated whereas 2.8% were over methylated in the BPD-DS group versus the control group (Figure 1b). The analysis of CpG sites within promoter regions revealed that five hypomethylated genes directly involved in insulin signaling (INSR, PIK3CG, PTEN, IRS1 and AKT2) exhibited the largest differences in methylation levels between BPD-DS and the control group (Table 2).
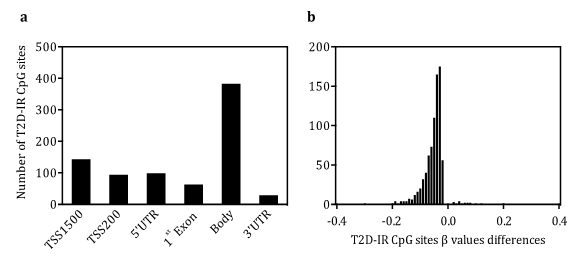
Figure 1 Differential methylation analysis a) Chromosomal distribution of T2D-IR CpG sites differentially methylated between BPD-DS and the control group (n=20/group). b) Histogram depicting β values differences in methylation levels between BPD-DS and control group (β>0 hypermethylation and β<0 hypomethylation in BPD-DS group). TSS: transcription start site; UTR: untranslated region. CpG: cytosine-phosphate-guanine.
Characteristics |
Control |
BPD-DS |
P |
Age (years) |
29.2 ± 3.3 |
41.0 ± 5.3 |
< 0.0001 |
BMI (kg/m2) |
45.8 ± 5.7 |
27.6 ± 4.8 |
< 0.0001 |
Blood pressure (mm Hg) |
|||
SBP |
130.0 ± 12.4 |
112.2 ± 9.5 |
< 0.0001 |
DBP |
81.3 ± 8.6 |
68.2 ± 8.9 |
< 0.005 |
Lipid profile |
|||
TC (mmol/l) |
4.68 ± 0.62 |
3.52 ± 0.49 |
< 0.0001 |
LDL-C (mmol/l) |
2.79 ± 0.58 |
1.68 ± 0.50 |
< 0.0001 |
HDL-C (mmol/l) |
1.22 ± 0.21 |
1.39 ± 0.25 |
0.023 |
TG (mmol/l) |
1.48 ± 0.68 |
0.97 ± 0.41 |
0.007 |
Glucose metabolism |
|||
Fasting glucose (mmol/l) |
5.2 ± 0.9 |
4.7 ± 0.3 |
0.038 |
Insulin (mU/mL) |
26.6 ± 23.9 |
3.1 ± 1.7 |
< 0.0001 |
HOMA-IR |
5.9 ± 5.0 |
0.7 ± 0.4 |
< 0.0001 |
Table 1 Characteristics of control and BPD-DS groups.
Data is expressed as mean ± SD (n=20/group). BPD-DS: Biliopancreatic Diversion with Duodenal Switch; P: T-test P-value; BMI: Body Mass Index; SBP: Systolic Blood Pressure; DBP: Diastolic Blood Pressure; TC: Total Cholesterol; LDL-C: Low-Density Lipoprotein Cholesterol; HDL-C: High-Density Lipoprotein Cholesterol; TG: Triglycerides; HOMA-IR: Homeostatic Model of Insulin Resistance
| Rank |
CpG ID |
Position |
UCSC Gene |
Localization |
P |
bDiff |
1 |
cg14900579 |
19:7294137 |
INSR |
TSS200 |
1.11 x 10-16 |
-0.300 |
2 |
cg08779777 |
7:106505772 |
PIK3CG |
TSS200 |
2.22x 10-16 |
-0.188 |
3 |
cg06947206 |
10:89623157 |
PTEN |
TSS200 |
3.33x 10-16 |
-0.100 |
4 |
cg19358349 |
10:89621871 |
PTEN |
TSS1500 |
3.17x 10-16 |
-0.095 |
5 |
cg11620807 |
2:227664353 |
IRS1 |
TSS1500 |
7.36x 10-16 |
-0.094 |
6 |
cg25333225 |
19:40791658 |
AKT2 |
TSS1500 |
1.80x 10-16 |
-0.090 |
7 |
cg14157042 |
20:58515396 |
PPP1R3D |
TSS200 |
2.22x 10-16 |
-0.080 |
8 |
cg04471409 |
16:67040954 |
RPS6KA2 |
TSS1500 |
1.46x 10-16 |
-0.079 |
9 |
cg20716209 |
17:40541270 |
STAT3 |
TSS1500 |
1.20x 10-16 |
-0.071 |
10 |
cg01678714 |
5:179780409 |
GFPT2 |
TSS200 |
1.20x 10-16 |
-0.062 |
Table 2 List of 10 most differentially methylated CpG sites between the BPD-DS and control group.
CpG ID: Cytosine-Phosphate-Guanine Probe Identification; Position: Chromosomal and Base Pair Position; UCSC Gene: University of California Santa Cruz Genome Browser Gene Mapping (GRCh37/hg19); Localization: CpG Localization Relative to UCSC gene; P: P-value of Differential Methylation t-test; bDiff: Average Methylation b Value Difference
Results from the present study show that most genes encoding proteins involved in T2D and IR pathways exhibited significant differences in methylation levels after BPD-DS when compared to a pre-surgery control group. Most of these loci were significantly hypomethylated, suggesting an effect of bariatric surgery on the epigenetic signature of genes involved in glucose homeostasis. Although there is no general consensus regarding the use of blood as surrogate of tissue-specific epigenetic changes,17 a global conservation of DNA methylation profiles has been previously identified between blood and adipose tissue in obesity.12,16 Accordingly, previous works have already reported differential hypomethylation induced by bariatric surgery in different tissues. Concretely, a recent study analyzing DNA methylation levels of isolated fat cells from SAT of women in a post-obese state also revealed a significant global hypomethylation,18 with an over-representation of genes involved in adipogenesis. Similar results were found in both SAT and VAT,19 as well as in skeletal muscle20 in women after bariatric surgery. Interestingly, differential methylation at promoter regions was associated in such studies with gene transcription alterations, and even with improvements in glucose homeostasis clinical parameters, such as insulin sensitivity.19,20 Thus, although further studies focused on the impact of post-bariatric hypo methylation in the transcriptional regulation of specific genes are still required, it is tempting to suggest that methylation changes found herein within T2D-IR-associated promoter loci may partly contribute to long-term effects of the BPD-DS on T2D resolution and improvements in IR.
Knowledge of epigenetic determinants predictive of obesity-related complications and metabolic improvements after bariatric surgery has considerable public health implications. Although further studies are still needed to establish a causal relationship between methylation changes in T2D-IR genes and metabolic outcomes after bariatric surgery, these epigenetic factors may be used as biomarkers to identify high-risk individuals that may be targeted for specific prevention and treatment programs, thereby reducing the burden of obesity and associated metabolic disturbances.
We thank the surgeons of the Department of Bariatric Surgery at the Quebec Heart and Lung Institute (Laurent Biertho, Simon Biron, Frédéric-Simon Hould, Stéfane Lebel, Odette Lescelleur, Simon Marceau) for their involvement in clinical care and participant recruitment. We express our gratitude to Suzy Laroche for help in sample and clinical information collection and Paule Marceau for participant recruitment, data management and project coordination. We acknowledge the contribution of the biobank staff of the IUCPQ-UL for sample management and the work of the McGill University and Génome Quebec Innovation Centre for gene methylation array processing. JTM has a postdoctoral position at the Institute of Nutrition and Functional Foods (Laval University). MCV is Canada Research Chair in Genomics Applied to Nutrition and Health. This study was supported by a grant-in-aid from the Heart and Stroke Foundation of Canada (G-14-0005824). AT receives research funding from Johnson & Johnson Medical Companies for scientific work on bariatric surgery.
Author declares that there is no conflict of interest.

©2017 de, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.