International Journal of
eISSN: 2573-2889


Research Article Volume 2 Issue 1
Department of Biotechnology, Panjab University, India
Correspondence: Jagdeep Kaur, Professor, Department of Biotechnology, BMS Block-1, Sector 25, Panjab University, Chandigarh 160014, India, Tel (0)0172-2534085, Fax 91-172-2541409
Received: November 10, 2016 | Published: January 23, 2017
Citation: Khurana J, Pratibha, Cameotra SS, et al. Studies on recombinant lipase production by E. Coli: effect of media and bacterial expression system optimization. Int J Mol Biol Open Access. 2017;2(1):17-23. DOI: 10.15406/ijmboa.2017.02.00008
Recombinant protein production is a vast field growing larger in each passing year due to continuous emergence of new host, vectors and techniques for cloning and expression. Earlier, a Bacillus lipase gene was cloned in pGEMT/ E. Coli DH5α with 12 units/ml lipase production. In the present investigation, attempt has been made to optimize the production of recombinant lipase by E. Coli by varying different parameters. Amongst the different strains of E. coli, M46 cells were observed to be the best host with 19units/ml lipase production. Addition of surfactant and changing media to nutrient broth lead to 2.8 fold and 1.3 fold respective increase in lipase production. Amongst different vectors, pET 28a vector showed the highest total lipase production but it proved toxic for the cells, while pQE30-UA vector gives 5 fold enhanced total lipase production with more stable expression. With the combination of optimized host, vector, surfactant and media, 18 fold increases in lipase production (214units/ml) was achieved.
Keywords: lipase production, coli, host, vector, enzyme activity, surfactant, promoter, optimisation
Lipases (triacylglycerol acylhydrolase, EC 3.1.1.3) are versatile enzymes with multiplexity of catalysis and therefore, constitute an important class of industrial enzymes in basic and applied research. They are part of the serine hydrolase family that acts on carboxylic ester bonds of tri-, di- and mono-glycerides and converts them into fatty acids and glycerol. According to recent surveys, by 2020 the lipase market is estimated to reach about $590.5 million, at a CAGR of 6.5% from 2015 to 2020.1 The potential of this enzyme as biocatalyst is literally boundless in many industries because of many characteristics like high stability in organic solvents, broad range of substrate specificity, high regio- and/or stereo-selectivity in catalysis and ability to act under mild conditions.2 Microbial lipases are the most useful ones as compared to the lipases from plant and animal origin because of high yield, ease of genetic alteration, broad range of catalytic activities, variety and tolerance to environmental conditions, regular supply due to absence of seasonal fluctuations and along with that fast growth rate of microbes in comparatively less expensive medium.3 The most preferred microbial sources used in the industry for production of lipases are bacterial and fungal sources. Among the extracellular lipase producing bacteria, the most important genus that have been exploited are Achromobacter,4 Alcaligenes, Arthrobacter, Bacillus,5 Burkholderia, Chromobacterium,6 Staphylococcus,7 Lactobacillus, Streptococcus,8 Micrococcus, Propionibacterium.
Several Bacillus lipases have been applied biotechnologically due to their versatility.9,10 Bacillus lipases have been isolated from various type of sources ranging from hot springs,11 to polluted areas in Port Dickson12 and salty soil.13 Due to such a wide range of origin Bacillus lipases exhibit different features including thermophilic,14 hyperthermophilic, psyrophilic,15 alkalophillic,16 acid thermo tolerant17 and organic solvent tolerance.18,19 Though several proteins are being produced at commercial scale by the native strain, but the production of recombinant proteins still poses a challenge in many cases. High production cost of any enzyme restricted its usage at large scale. Optimization to increase the lipase production, without increasing the cost, is very important. Few strategies that can be adopted to reach the goal are changing expression host cells and expression vector/promoter, optimization of media composition, addition of some components to growth media, optimization of culture conditions etc. Many Bacillus lipases had been cloned, expressed, purified and characterised to an extent until date.20‒22 Various studies has been carried out to optimise the production of recombinant proteins in E. Coli by changing media composition.23‒25 Some studies have also been carried out to optimise the expression system and host cells for the production of recombinant proteins in heterologous system.11, 26, 27
A lipase gene had been cloned from Bacillus sp. by Khurana et al.,28 This recombinant protein demonstrated activity at broad pH and optimum activity at 35°C. In addition, this enzyme showed activity at low temperature and tolerance to various organic solvents. The enzyme might be a good candidate for industrial applications. Therefore, in the current investigation, an effort was made to optimize the production of this recombinant Bacillus lipase in E. Coli with respect to host strain, surfactant, media and different expression vectors in shake flask experiments.
Chemicals, bacterial strains and plasmids
All the chemicals and reagents used were of analytical grade and purchased from MERCK (Germany) and Hi Media (India). Lipase substrate para-nitrophenyl laurate (C12) was purchased from Sigma Chemicals Co. (USA). All the E. Coli host cells used in the study were procured from MTCC, IMTECH, and Chandigarh, India. The plasmids used in the study were purchased from Promega, USA (pGEMT) and Qiagen, Germany (pQE-30 UA and pET28a). Antibiotics, IPTG and X-gal were purchased from Hi Media (India).
Growth conditions
The E. Coli cells were grown in Luria–Bertani medium (LB), pH 7.4 at 37°C, 180rpm. The micro-organisms were inoculated (1%) in production medium (LB, 2% w/v) containing suitable antibiotic. The cells were induced with 0.05 mM IPTG at start of the culture.
Enzyme assay
The enzyme activity was determined according to the modified method of Singh et al.29 To 700µl of 50mM phosphate buffer (pH 8.0), 100µl of sodium deoxycholate (10mM), 100µl of enzyme and 100µl of 2mM p-nitrophenyl laurate were added. The reaction was carried out at 37°C for 10min, after which 250µl of 0.1 M Na2CO3 was added. The mixture was centrifuged for 5min and the activity was determined by measuring absorbance at 420 nm. One unit of enzyme activity is defined as the amount of enzyme, which liberates 1µmole of p-nitrophenol from pNP-laurate per min under standard assay conditions. All the experiments were performed in triplicate and mean and standard deviations were calculated.
Host dependent lipase production
A range of E. Coli host cells (M-46, X42, DH5α, JM109, M-15, M390, M1668 and BL21) were checked for lipase production. Competent cells were prepared of all the strains and transformed with pGEMT-lip plasmid by heat shock method at 42°C for 90sec. Supernatant and pellet of the overnight grown culture of transformed cells were assayed for the lipase production using standard assay condition and relative enzyme activity was calculated. DH5 α cells transformed with pGEMT-lip plasmid were served as the control.
Effect of different nutritional media on lipase production
pGEMT lip/M46 cells were grown in four nutritional media i.e. nutrient broth (NB), Luria-bertani broth (LB), YT media and SOB media for 24h. Supernatants of cultures were assayed for lipase production to check the effect of these nutritional media on lipase production.
Effect of surfactant on lipase production
The surfactant used in this study was a rhamno lipid. pGEMT-lip / M46 cells were grown in culture media (2% w/v LB) with varying concentration (0.05, 0.1, 0.5, 1 and 1.5mg/ml) of surfactant for 24h. Assay for lipase activity was carried out at an interval of 3 h and relatively compared with the control media (no surfactant added). Effect of time of incubation with optimum concencentartion. of surfactant on enzyme activity was also assayed. Also, the effect of surfactant on growth of culture was checked by taking absorbance at 600nm.
Effect of different vectors on lipase production
The effect of various expression vector on lipase production was determined by cloning lipase gene in different vector. The pGEMT-lip plasmid was used as a template for amplification and the primers used for the cloning were mentioned in Table 1. Amplification protocol and conditions were same as in Khurana et al.28 Amplified product was ligated directly with pQE30UA vector. But, for cloning in pET28a vector, both the vector and amplified product were digested with BamHI and Hind III restriction enzymes and then ligated with each other. The ligation mixtures were transformed into E. Coli. M15 cells and E. Coli. BL21 (DE3) cells respectively. Recombinant clones were screened on LB agar plates supplemented with selective antibiotic and further orientation was confirmed by colony PCR and sequencing. Lipase production enzyme was carried out and enzyme activity was calculated for both extracellular and intracellular fraction.
Vector |
|
Sequence |
pQE30 UA |
Forward |
GATCGCCCATGGGTGAAAAAAGTACTT |
Reverse |
GATCGCGGATCCTTAATTTGTATTGAG |
|
pET28c |
Forward |
GGATCCATGGTGAAAAAAGTACTTATGG |
Reverse |
AAGCTTTTAATTTGTATTGAGGCCTCC |
Table 1 Sequence of primers used for cloning of lip gene in expression vectors
Initially, we had cloned a Bacillus lipase (lip) gene in pGEMT vector/ E. Coli DH5α cells. In spite of the fact, that we had used cloning vector, there was some level of expression (12 Units/ml) of recombinant lipase. To further, enhance the production of lipase, different strategies of over-expression such as varying host, vector, production media and addition of surfactant were employed in the present investigation.
Effect of different host cells on lipase production
To determine the effect of different E. Coli host cells on recombinant lipase production, a wide range of host cells were transformed with pGEMT vector carrying lipase gene (pGEMT-lip). Host cells of E. Coli M46 and X42 demonstrated the best lipase production in comparison with other host strains on tributyrin agar plate. Further, these host cells were grown in liquid media to determine the extracellular and intracellular lipase production (Table 2). Amount of lipase in intracellular fraction was nearly equal in M46 and X42 host cells followed by JM109 cells. However, total lipase production and extracellular production (19 units/ml and 15 units/ml respectively) was observed to be maximum in M46 cells. However, the growth rate (data not shown) and relative distribution of lipase in extracellular and intracellular fraction was observed to be nearly same in all the host strains (Table 2).
Host cells |
Lipase Production (U/10 ml) |
Relative Lipase Production (activity in %) |
|||
|---|---|---|---|---|---|
|
Extracellular |
Intracellular |
Total |
Extracellular |
Intracellular |
DH5α |
95+3 |
24 +4.1 |
119+ 7 |
79+3 |
21+3.3 |
M-46 |
150+3 |
40 +1 |
190+5 |
79+2.8 |
21+22 |
X42 |
134+4 |
40+ 3 |
174+7 |
73 +2 |
27+3.8 |
JM109 |
121+2 |
34+ 2 |
155+ 4 |
78+ 3.1 |
22+5.8 |
M-15 |
90+3.1 |
25+ 3.3 |
115+7 |
78+2.5 |
22+1.1 |
M390 |
102+2.2 |
27+ 2 |
129+6 |
79+2.3 |
21+6.1 |
M1668 |
105+2.6 |
30+ 4 |
135 +7 |
78+3 |
22+3 |
BL21 |
88+ 3.2 |
25+ 1.8 |
113 +5 |
77+3 |
23+4.2 |
Table 2 Effect of different host cells on recombinant lipase production
Effect of different media on lipase production
Since culture media can significantly control the growth and performance of microbial processes and hence the productions of desired compound/protein, four different production media were checked for their effect on lipase production in M 46 host cells. Supernatant was assayed for enzyme activity in extracellular fraction. In minimal media there was negligible production of lipase. However, maximum lipase production was observed in the nutrient broth which was almost 1.3 folds higher than in Luria Broth (Figure 1).
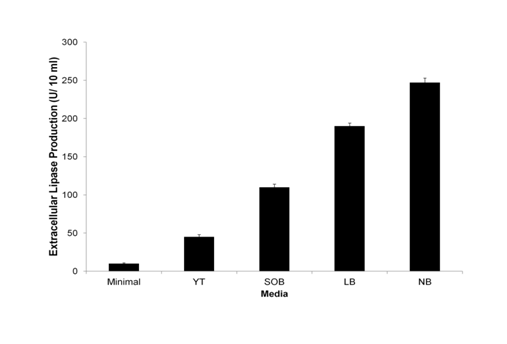
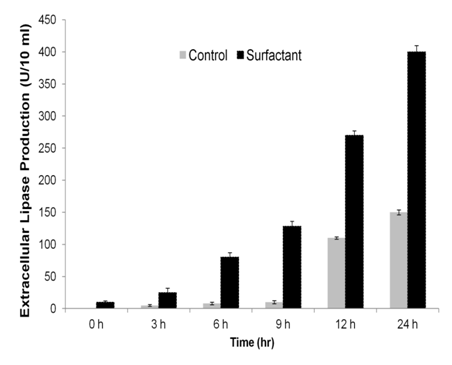
Effect of surfactant on lipase production
Surfactants are reported to play important role in overall lipase activity. Therefore, different concentrations of surfactant were added in the culture medium and were incubated at 37°C for 24h and lipase production was determined after every 3h. The culture without surfactant started secreting lipase after 9h of incubation (Figure 2) while the lipase activity was detected even after 3h of incubation in culture with surfactant at 0.1-0.5mg/ml concentration. However, further increase in the concentration of surfactant (at 1.0 and 1.5mg/ml) lead to decrease in the production of lipase.Out of different concentrations of surfactant, 0.5mg surfactant/ml displayed maximum relative lipase production. Therefore, 0.5mg/ml surfactant was selected for further studies. The fold enhancement of lipase production was studied with 0.5mg surfactant/ml of media at different time intervals (Figure 3). The culture without surfactant served as control and it was observed that there was increase in lipase activity to 21 folds after 6h and 14 folds after 9h of growth of culture with surfactant. After 12 and 24h of incubation lipase activity was almost 2.8times of control (Figure 3a). However, surfactant did not affect the growth of host carrying pGEMT-lip (Figure 3b). This nullifies the possibility that difference in lipase production was due to difference in growth of bacteria.
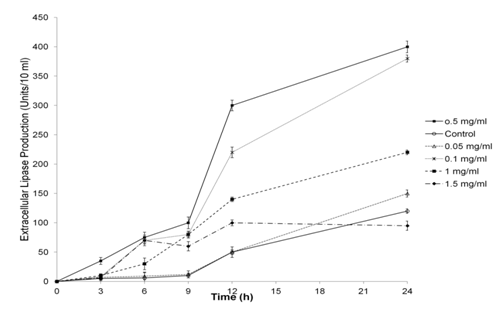
Effect of different promoters on lipase production
In the present investigation, initially the lipase production was being observed in pGEMT /E. Coli DH5α with 12units/ml. Since it is cloning vector, expression was observed to be less, variable and unstable Table 3& Figure 4. Then, we tried two expression vectors i.e pQE30 UA and pET28a for expression of genes. In pET28a vector, lipase production was under tight regulation of T7 promoter in E. Coli BL21 (DE3) cells. No extracellular activity was detected when lip gene was induced by 0.1mM IPTG at 37oC for 3h (data not shown). However, lipase production was observed after overnight induction by 0.05mM IPTG at 30oC. Extracellular and intracellular lipase production was found to be 56U/ml and 96U/ml respectively (Table 3), which very much higher than total production in pGEMT /E. Coli DH5α. But, unfortunately, the growth of micro-organism was severely impaired (Figure 4) even when pET-lip was induced at very low temperature.
|
Enzyme production(U/10 ml) |
Relative lipase activity (%) |
||||
|---|---|---|---|---|---|---|
Vector |
Extracellular |
Intracellular |
No sub culturing |
First |
Second |
Third |
pGEMT |
95+3 |
25+4 |
100 |
69+2.4 |
48+3.4 |
25+2.8 |
pQE30 |
464+3 |
136+2 |
100 |
92+3 |
89+3 |
85+3 |
pET28a |
560+6 |
960+8 |
100 |
Growth impaired |
- |
- |
Table 3 Effect of different Vectors on lipase production
Further, the expression of lip gene under the influence of T5 promoter/pQE 30UA in M-15 host cells was induced by 0.05mM IPTG overnight at 30oC. In comparison to lipase production in pET28a vector, 32U/ml extracellular lipase activities was observed in pQE 30UA vector (Table 3). However, production of lipase was stable at 30oC. Moreover the growth of cells was not hampered (Figure 4) even after 3 subculture. The stable lipase production was further confirmed by three passages followed by induction by IPTG (Table 3). The pGEMT clone demonstrated nearly 75% loss of enzyme activity after 3rd passage while in pET28a clone the bacterial growth was highly impaired after the first passage. The lipase expression in pQE 30UA vector was stable even after 3rd passage.
Further different combination were tried to enhance the production of lipase. Maximum lipase production was observed in M 46 cells therefore pQE-lip was transferred into E. Coli M-46. M-15 cells have pREP 4 plasmid in their system. Therefore, pREP 4 plasmid was isolated from E. Coli M15 host cell and M 46 cells were first transformed with pREP4 vector followed by pQE-lip. Cultures were grown in nutrient broth at 37°C until absorbance reaches 0.4-0.6. Cultures were induced with 0.05mM IPTG incubated for overnight at 30°C. Enzyme activity was measured in both supernatant and pellet. In comparison to M15/pQE-lip, M46/pQE-lip demonstrated nearly 1.5 folds enhancement in extra-cellular enzyme activity (Table 4).
Host cells |
Vector |
Lipase activity (units/10 ml) |
|
|---|---|---|---|
|
|
Extracellular |
Intracellular |
M 15 |
pQE30 |
472+13 |
144+ 14 |
M 46 |
pQE30 |
703+30 |
168+23 |
Table 4 Effect of T5 promoter on lipase production in different host cells.
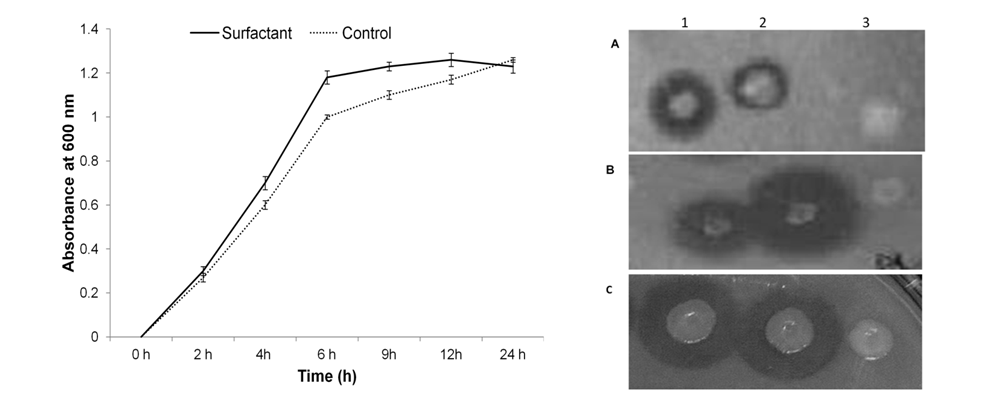
The different combination of production media shows the enhancement in lipase activity of M46/pQE-lip. The maximum total lipase production (214 units/ml) and extracellular lipase production (192units/ml) was observed with pQE+M46+NB+ Surfactant (Table 5). Relative distribution of total lipase in extracellular and intracellular fraction also vary with the change in production media. With the addition of surfactant to LB media ratio of extracellular to intracellular lipase changes from 5:1 to 7.5:1, while addition of surfactant in NB media, ratio of extracellular to intracellular lipase changes from 5:1 to 9:1. Therefore, the expression of lipase gene under the T5 promoter was enhanced by 3.5 folds by choosing , E. Coli M46 strain as host, NB media and surfactant than E. Coli M15 strain as host and LB media. This total lipase production (214U/ml) was nearly 18-20 folds higher than the initial lipase produced by pGEMT-lip (12/ml) in DH5α.
Combination |
Lipase production (Units/10ml) |
||
|---|---|---|---|
|
Extracellular |
Intracellular |
Total |
pQE+M15+LB |
483+6 |
125+8 |
608+14 |
pQE+M46+LB |
705+11 |
148+11 |
854+20 |
pQE+M46+NB |
935+15 |
171+4 |
1106+21 |
pQE+M46+Surfactant+LB |
1768+21 |
236+3 |
2004+22 |
pQE+M46+NB+Surfactant |
1923+15 |
223+ 9 |
2146+22 |
Table 5 Lipase production in different combinations of production media
Protein over-expression in bacteria is still the easiest, cheapest and therefore preferred way to obtain large amount of proteins. Despite several new hosts developed in the recent years, bacterial cells specially the E. Coli remain the preferred system for the production of recombinant proteins because of the ease of its genetic manipulation, the availability of various host/vector systems, and its fast growth.30 There are several issues relating to gene expression in E. Coli such as protein solubility, protein degradation by proteases, chaperones, the complexity of in vivo aggregation and cell death that needed lots of attention. It has been reported that E. Coli envelope is able to sense and actively react to protein over-expression in the cytoplasm.31 However, it reacts to protein over-expression in the cytoplasm in a generalized and non-specific way by improving the traffic of small solutes, i.e. nutrient uptake as well as elicits changes in the membrane proteins. Over-expressed proteins are often produced in the form of inclusion bodies,32 from which biologically active proteins can only be recovered by complicated and costly denaturation and refolding processes. A variety of techniques have been developed to solve these problems, including the use of different promoters to regulate the level of expression, the use of different host strains, co-expression of chaperones, changing media composition and reduction of culture temperature etc.33,34 Therefore, a study on effect of different E. Coli host, promoters, surfactant and production media on over-expression of lipase was carried out in the present investigation.
First, we tried different E. Coli host strains because there are various E. Coli host strains that have been developed to complement different requirements in gene cloning and expression. The strain or genetic background for recombinant expression of protein in heterologous expression is highly important. Since, expression strains should maintain the expression plasmid stably, should be deficient in harmful natural proteases and confer the genetic elements relevant to the expression system. The cloned lipase had its original signal sequence from Bacillus sp. We recently demonstrated that the signal sequence of this lipase is not processed and the extracellular protein retains the signal sequence.28 In the present study, though the relative amount of lipase present in supernatant and pellet is nearly same in all the host cells, the overall lipase production was varying in different strains with the highest in E. Coli M46 cells. It showed that E. Coli M46 cells might be the best complemented with the cloned gene of interest. Vincentellli et al, 2011 used four different strains E. Coli BL21 (DE3) pLys S (B), Ros (DE3) pLys S (R) and Ori (DE3) pLys S (O), for expression of 96 proteins along with 3 different temperature and 3 different culture media. They found out the optimised conditions (out of 12 conditions) for each protein using auotomated dot plot method.34
The composition of the cell growth medium may also have significant metabolic effects on both the cells growth and protein production. There are three types of media defined, complex and semi-defined. Defined media are generally used to attain high cell density, as the nutrient concentrations are well-known and can be controlled during culture. However, complex or semi-defined media are sometimes necessary to boost product formation.35 We had selected four complex media i.e Luria Broth, Nutrient Broth, YT media and SOB, with minimal media as control to study their effect on lipase production from our recombinant culture. The Nutrient Broth showed maximum extracellular lipase production which was almost 1.3 folds higher than in Luria Broth. Vincentelli et al.34 reported that culture medium composition (SB; 2YT; TB) was not a major determinant of protein solubility for both prokaryotic and eukaryotic targets as solubility was the same per cell. So more the biomass the more will be the amount of protein produced.34 Like most of Bacillus lipases, this lipase also formed aggregates at high protein concentration. Reduction in formation of aggregates could increase the amount of available soluble protein. Numerous solution additives had been proved useful for this goal. It had been reported that the non-detergent surfactants are more favourable for aggregation suppression than the detergents.36 Therefore, we add surfactant to the growth media and observed 2.8 fold enhancements in enzyme activity when compared with production without surfactant. Surfactant might be helping lipase to stay in soluble form even at high concentration, as usually proteins are most soluble in the native folded state in aqueous solution. When the structure deviates from the native state, proteins tend to aggregate. Similarly37 found a two-fold increased recovery of Aspergillus niger lipase when Triton X-100 was used as surfactant. Addition of surfactant induced the lipase production in lipolytic fungi with the maximum induction (4 fold) with Tween 80.38
Even after the above optimisations, we observed a variable and unstable expression due to pGEMT vector. Hence, we tried to change the vector i.e. pET28a and pQE 30UA vector. To reduce the toxicity, lipase activities of different expression system were carried out using the original leader sequence. Because, the prolipase was reported to be less toxic to its host than the mature lipase.39 The lipase was produced as soluble protein in both the expression systems. Though the lipase production was higher in pET/BL 21 system than pQE/M15, however, the lipase activities of the expression system/ expression level under the control of T7 promoter was not very stable. The high expression of lipase in E. Coli BL21 DE3 seemed to be toxic for the cell as despite the big zone of clearance around the place where the colony was patched, no bacterial growth was observed. It has been reported that in E. Coli heterologous protein expression of certain proteins are toxic even when levels of expression are very low.40 Also, basal transcription of protein may introduce a cellular stress situation and thereby selects for loss of recombinant plasmid i.e. unstable expression.41 These might be the reason for toxicity and instability of protein expression in the pET/BL21 system. However, the lipase cloned in pQE30-UA vector demonstrated stable expression of lipase gene. The pQE-30UA vector possess a phage T5 inducible promoter and two strong transcriptional terminators t0 from phage lambda and T1 from rrnB operon of E. Coli to prevent read through transcription and ensure stability of expression. Moreover, M15 cells harbours pREP4 plasmid that encodes for lac repressor which ensures that induction will start after the addition of inducer (IPTG) only and not prematurely, thus less toxic for the host cell.42 For these reasons we transformed M46 cells with pREP4 vector while checking expression in the E. Coli M46 cells. On contrary11 demonstrated 11 fold higher lipase production in pET-51b/ BL21 (DE3) pLysS, in comparison to pQE-30/ M15(pREP4) system. Finally, we tried different combination of host (M46/M15), media (LB/NB) and surfactant to find out the best combination. At the end, we could enhance the lipase production drastically (18-20 fold) by using the optimized host (M46), expression vector (pQE-30UA), media (Nutrient broth) and surfactant.
In the present study, we optimized the conditions for our recombinant lipase production in E. Coli. The total enzyme production was observed to be enhanced with the change of host/media and addition of surfactant. But, as overproduction of lipase is sometimes detrimental for the survival of host E. Coli cells this enhancement should not affect the bacterial growth. Therefore, we changed the promoter to T5 to provide stable over expression of lipase. With the above mentioned combination, we could enhance the lipase production by 18-20 folds by choosing proper host cell, expression vector, production media and surfactant in vectors in shake flask experiments. Further, we will try to scale up the production of this enzyme in fermenter.
None.
Author declares that there is no conflict of interest.

©2017 Khurana, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.