International Journal of
eISSN: 2573-2889


Research Article Volume 2 Issue 1
1Department of Zoology, University of Allahabad, India
2Department of Zoology, Nehru Gram Bharati University, India
Correspondence: Sandeep K Malhotra, Department of Zoology, University of Allahabad, India
Received: October 30, 2016 | Published: January 11, 2017
Citation: Jaiswal N, Malhotra SK. Molecular characterization of host-specific raphidascaridoid worms from the gangetic garfish (teleostomi: belonidae) in India. Int J Mol Biol Open Access. 2017;2(1):1-15. DOI: 10.15406/ijmboa.2017.02.00007
The 18S rDNA gene analysis with Internal Transcribed Spacers 1 and 2, and Mit. coi gene analyses of 12 sequences from the host-specific nematodes parasitizing liver of garfish, Xenentodon cancila (Belonidae) revealed their monophyletic characterization. The purification of PCR products was done using Centri-Sep spin columns (Princeton Separations, Adelphia, New Jersey, USA) and sequenced by ABI 3730 auto sequencer Applied Biosystems. Their genetic heterogeneity on the basis of comparable sequences from gnathostomatoid genus, Gnathostoma, 16 anisakid and ascaridoid genera assisted these to be ascribed to a newer genus, Indospinezia multispinatum gen. et sp. n. under a newly erected tribe Indospineziinea tribe n. under family Goeziinae, as the morphological differentiation distinctly segregated their taxonomic identity. The fish host, X. Cancila acted as the common intermediate as well as definitive host in the Gangetic riverine ecosystem at Allahabad, India. Though the newer worms had typical gnathostomatoid (fish spirurins) characters, yet the single-toothed unidentate spines, cervical sacs, and the number of caudal papillae, that comprised pre-cloacal foliate as well as ‘sunflower’ papillae, differentiated them from the spirurins. The post-labial double-ridged cuticular collar at the base of head, and an altogether different sclerotized oral lid-plate distinguished these worms from genus Goezia as well as spirurins. Therefore, the new genus, has been accommodated under a newly raised tribe, Indospineziinea tribe n.
Keywords: molecular characterization, Indospinezia multispinatum et sp.n, raphidascaridoid; indospineziinea tribe n, anisakid
Anisakidosis from the parasitized fish in fresh water riverine ecosystems has emerged in recent years as a serious cause of concern to human beings. A great variety of human infections of anisakidosis have been compiled in earlier investigations.1 The irritation of the oesophagus and stomach, vomiting, nausea, diarrhea leading to severe epigastric and abdominal pain were the clinical symptoms on record. The chief causative agent to elicit severe clinical symptoms in human beings were the third stage larvae of Contracaecum termed contracaecosis,1 Pseudoterranova termed pseudoterranovosis2 and Anisakis spp. termed anisakiosis.3The mechanism of penetration by P. Decipiens & C. Osculatum into the mucosa of gastro-intestinal tract has also been investigated in recent years.4 The state of knowledge about Anisakis (Dujardin), Contracaecum (Railliet and Henry), and Pseudoterranova (Mozgovoi) causing widespread painful infections in the digestive tract in human beings through consumption of semi-cooked fisheries products was reviewed recently.5 Uneven distribution of larvae of various species of Anisakis larvae was also concluded by application of molecular studies.6 Resultantly the distribution of A. Simplex (s.s.) was found restricted to the northern hemisphere, while A. Typica, though yet to be proved pathogenic, was found distributed only in the tropical regions.7 The first report of human anisakidosis by consumption of South Australian Mackerel was recorded from locally caught fish in Australia.8
The life cycles of nematodes have contributed significant information to formulate earlier schemes of their classification by eminent taxonomists, which have been adequately reviewed.9 The significance of varied patterns in the life cycle of a nematode in the evolutionary events was highlighted by several earlier workers.10,11 In recent years, notable attempts have been made by taxonomists world over to develop phylogenetic hypotheses for cracking the mystery of interrelationships among ascaridoid and other taxa.12‒14 G. Bangladeshi was raised on the premise of the similarity of its life cycle with members of subfamily Raphidascaridinae on the basis of morphology of its juveniles, but without recovery of its developmental stages, particularly second or third stage larvae.15 Although the newer worms possessed several features similar to genus Goezia, like ‘cactus’ tail bearing ‘sunflower papillae’, yet the report of a larval tooth in the second stage larvae,16 a post-labial collarette, oesophagus with an anterior muscular and a posterior glandular part and the genetic differentiation from Goezia strengthened their position as a separate taxon within family Raphidascarididae.
The characterization of ascaridoid nematodes in fish with comparative account of morphology of worms have been discussed by several eminent workers.17,18 There has been no report on the same individual fish serving as intermediate as well as definitive host of a Raphidascaridoid nematode from freshwater fish hosts in India. The distinctive taxonomic status of family Anisakidae, comprising subfamilies Anisakinae and Contracaecinae, separate from Raphidascarididae, has already been recognized.19 The transversions-weighted bootstrap tree published earlier13 did not recover the grouping of Goezia with anisakids, but Goezia was a part of Raphidascarididae with higher (99%) bootstrap support in the combined evidence tree. Accordingly, both the most parsimonious explanation of the combined data and statistical evaluation refuted inclusion of Goezia with the anisakids.13 Therefore, the author’s contention to include Goezia under Raphidascarididae, along with Raphidascaridoides in the revised key to family Raphidascarididae has gained ground. An additional feature of occurrence of larvae in identical habitat, i.e. liver of fish, was similar between the new taxon and Gnathostoma spinigerum which brought the former closer to fish spirurines. The fish host of the newer genus was, however, a freshwater omnivorous column feeder, X. Cancila in River Ganges at Allahabad, contrary to the predominantly marine anadromous fish host Tenualosa ilisha (Clupeidae), of G. Bangladeshi, in Bangladesh. The latter migrated from Indian Ocean to River Ganges, for spawning purposes, and survived in the fresh water tributaries of this River to enter into Bangladesh, before finally passing life in the Ocean. The relationships of Spirurina inferred from SSU rRNA gene sequences demonstrated17 significant differences between Gnathostomatidae and other Spiruromorpha at the molecular level, whereas their strong affinity to Ascaromorpha was peculiar. The family Goeziinae is proposed to be split into two newly raised tribes, firstly Indospineziinea Tribe n. and Goeziinea Tribe n. to accommodate the newer genus Indospinezia multispinatum gen et sp. n. under the former tribe.
Collection and examination of fish for parasites
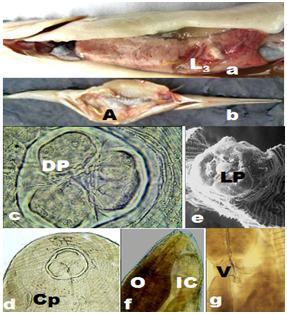
The LSID for this publication is lsid: zoobank.org: act: 7E42164F-A236-4EC1-92CB-74FAE30721F2. The investigations comprised parasitological examination of 2081 Xenentodon cancila, during 2008-2012, out of which 34 of 497 during 2008-2009, 91 of 566 during 2009-2010, 272 of 588 during 2010-2011, and 80 of 430 fish during 2011-2012 were found to be infected. The fresh bottom dwelling fish were collected from the fishermen at River Ganges, Yamuna and Allahabad in Uttar Pradesh, India. The second stage larvae, early third stage larvae, advanced third stage larvae heretofore called as AdvL3 Figure 1a and mature worms (Figure1b &4i) were frequently encountered in a cavity within the liver tissue of infected fish under natural conditions. The mature worms or larvae never occurred in GI tract nor they formed nodules in its wall, but instead favoured hepatic dwellings although the eggs and various other developmental stages, including second stage larvae were recovered by pepsin digestion method20 treatment of muscles and tissues of the fish.
Adult nematodes and AdvL3 were killed in lukewarm water; washed thoroughly in 0.85% normal saline, and fixed in Berland’s solution. A small piece of the mid-body of each of the 11 individual nematodes and one AdvL3 were removed with a scalpel and preserved in 100% ethanol for molecular analysis. The remainders of the roundworms were processed for morphological examination.14 Five mature roundworms and one AdvL3 were fixed in Glutaraldehyde (2.5% in 0.1M phosphate buffer) for the Scanning Electron Microscopy (SEM) and processed after rehydration.14 SEM analysis was performed on Jeol JSM 6510LV at the University Sophisticated Instrument Facility (USIF), Aligarh Muslim University, Aligarh, India. Adult nematodes and their larvae isolated from the liver of the definitive host fish, X. Cancila, were identified to genus and species levels based on the available keys and descriptions.18,21‒25 Drawings were made with the aid of a drawing tube and measurements were made directly with an eyepiece micrometer. Measurements of roundworms were recorded in µm and expressed as range, followed by mean in parentheses, unless otherwise stated. Photomicrographs were taken using Motic research microscope with Biovis Image Analysis software. Specimens have been deposited in collections of the Zoological Survey of India, Dehradun, India.
Molecular analyses
The total genomic DNA was extracted by standard phenol-chloroform method26 from individual worm, followed by an overnight digestion with proteinase-K at 370C, alcohol precipitation and washing with 70% ethanol or, using Qi Aamp Tissue Kit (Qiagen). The pellet of DNA was suspended with 50µl of Tris-EDTA (TE) buffer. The concentration of isolated DNA was estimated using a UV spectrophotometer. The DNA was diluted to get a final concentration of 50 ng /µl. The mitochondrial DNA (mt DNA) Cytochrome C Oxidase subunit I (coi) and small subunit ribosomal RNA (18S rRNA) genes were amplified by Polymerase Chain Reaction (PCR) technique27 using “BIORAD Thermo Cycler”, and primers.14 The DNA sequences were aligned for phylogenetic analysis using the Clustal W computer program28 and DNA sequences were edited in BioEdit.29 The evolutionary distances were computed by Kimura’s two parameter method.30 The neighbor joining tree31 was generated using Molecular Evolutionary Genetics Analysis (MEGA) software version 5.32,33 The tree was evaluated using the bootstrap test values34 based on 1000 replications.
The rDNA region comprising the ITS1 and ITS2 sequences was amplified with primers, SS1, 5’-GTTTCCGTAGGTGAACCTGCG-3’; and NC13R, 5’-GCTGCGTTCTTCATCGAT-3’ for ITS1 gene and SS2, 5’-TTGCAGACACATTGAGCACT-3’; and NC2, 5’-TTAGTTTCTTTTCCTCCGCT-3’ for ITS2 gene.33 Each PCR reaction was performed under the following conditions: after initial denaturation at 940C for 5min., 35 cycles of 940C for 30s (denaturation), 500C for 30s (annealing), 720C for 30s (extension), followed by a final extension at 720C for 8min. An aliquot (4 ml) of each amplicon was examined on a 1.5% w/v agarose gel, stained with GelRedTM and photographed using a BIOVIS gel documentation system. The purification of products was done again using Centri-Sep spin columns (Princeton Separations, Adelphia, New Jersey, USA) and sequenced by ABI 3730 auto sequencer (Applied Biosystems). The trees were out group-rooted using the sequences from gnathostomatoid genus, Gnathostoma, 16 anisakid and ascaridoid genera, as listed in Table 4 with whom the new genus has been genetically compared. Partial Mit. coi sequences with 18S rDNA, ITS1 and ITS2 gene sequences34 of specimens in the present study were compared for homology with sequences present in the Genbank of the National Center for Biotechnology Information (NCBI) by using BLAST (Basic Local Alignment Search Tool).35
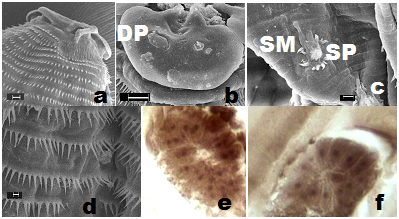
The roundworms were sturdy, cylindrical and light yellowish-brown in colour with typical anisakid ventriculus at the oesophageo-intestinal junction (Figure 1g). Female worms were distinctly larger and stouter than males. The body of larva grew enormously in size, and measured even longer than that of mature female worms. The body was enclosed in a cuticular envelope with a marked aspinose cephalic region with three lips Figures 1c&2a. The anterior end was distinctly demarcated by an elevated post-labial double-ridged collarette Figures 1c,2a&2b at the base of the head, from rest of the body, beyond which the spiny annulations occupied the whole body. The inner margin of the three shallow lips comprised two inwardly projected arms, each with a knob-like terminal end, that were equidistant from the triangular margins of the sclerotized lid-plate atop sclerotized stoma orally Figure 1e. The pedunculate base of each of the three lips was curved inwardly to merge on the side walls of the vestibule, while the pair of inwardly projected arms, with their distal end hanging free, emerged from each of the three lips at the level of curvature of pedunculate base. The three-cornered dorso-lateral anchorage of the lid-plate comprised a marginally ridged structure that encompassed a shallow groove in which individual terminal end of the lid-plate fit tight. This lid-plate with smoother external surface appeared as an adaptive device, and was designed as a filter during feeding, to suit site-specific location of nematodes in softer liver tissues. It had an apparent upward-downward slide, while the edges remained loosely affixed in the grooved corners of anchorage zone. This ensured that there was minimal erosion while sucking liquid from the available space at the margins of the vestibule, beneath the lid-plate in the cavity specially carved by the parasite on the surface of hepatic tissue. This enhanced the functional significance of the structurally modified elements in the oral morphology of roundworms. Of the three lips in mature male, female as well as AdvL3, one dorsal lip comprised two double papillae occupying lateral positions, closer to the margins of the twin-lobed projections oriented towards vestibule with a single amphid. Each of the two ventrolaterals (Figure 2a&3b) comprised one double papilla, one single papilla and a lateral amphid. A pair of deirids (Figure 4a) were present between 25th and 28th row of spines, and two unpaired dome-shaped papilla between 15th and 18th row of spines, and at 20th row of spines each. Nerve ring was located between 7th and 9th rows of spines in the fore body. Excretory pore opens latero-ventrally, located between 21st and 23rd rows of cuticular spines. Cervical sacs two pairs extended upto 24-42nd rows of spines. Oesophagus was divided into two parts Figure 1f-an anterior muscular and a posterior glandular. The hinder extremity of the oesophagus Figure 1g was followed by a prominent bulbar ventriculus, from which emerged ventricular appendix Figure 5b that was split into an elongated, bitubular structure, and an anteriorly directed intestinal caecum of globular shape usually, and overlapped major part of posterior glandular part of oesophagus. The transverse section of the oesophageal glands of AdvL3 and adult worms revealed presence of one to three nuclei Figures 3e,3f. Tail conical with digitate mucronate at the terminal end was covered with minute rows of spines till the posterior extremity. Caudal alae were prominent Figures 2h. Various measurements of different body organs of mature worms of the proposed new genus are summarized in Table 1.
Characters |
Male |
Female |
Body L |
5916–14269 (11050) |
11140–27080 (24161) |
W |
268–1377 (814) |
882–3978 (1889) |
Head L |
40– 81 (58) |
72–177 (108) |
W |
162–266 (219) |
265–390 (322) |
Lips L |
68–85 (77) |
68–108 (83) |
W |
102–164 (128) |
117–162 (135) |
Buccal cavity L |
19–31 (32) |
78–108 (89) |
W |
27–135 (82) |
39–315 (225) |
Glandular oesophagus L |
367–562 (473) |
475–1188 (859) |
W |
156–390 (219) |
198–864 (456) |
Muscular oesophagus L |
441–702 (690) |
810–1786 (1168) |
W |
138–415 (369) |
450–1206 (778) |
Oesophagus L |
659–2160 (1331) |
1800–2286 (1809) |
W |
176–938 (414) |
252–1224 (572) |
Oesophagus/body length ratio |
1:7–1:18 (1:12) |
1:7–1:8 (1:8) |
Cervical glands L |
238–362 (250) |
209–391 (380) |
W |
17–29 (27) |
21–44 (38) |
Ventriculus L |
88–187 (167) |
94–515 (339) |
W |
88–288 (226) |
221–972 (434.3) |
Ventricular appendix L |
1733–4500 (3146) |
2124–4950 (3489) |
W |
178–324 (238) |
100–416 (303) |
Ratio of Oesophagus/Ventricular Appendix L |
1:1–1:2 (1:2) |
1:1–1:3 (1:2) |
Intestine W |
97–215 (142) |
153–589 (350) |
Intestinal caecum L |
774–1116 (1000) |
1044–2556 (1525) |
W |
90–333 (198) |
90–297 (201) |
Distance of vulva–a. from anterior end |
- |
4977–7290 (6256) |
b. from posterior end |
- |
8028–11206 (10008) |
Eggs L |
- |
23–43 (33) |
W |
- |
35–59 (47) |
Left spicule L |
371–488 (325) |
- |
W |
16–55 (34) |
- |
Right spicule: L |
320–382 (356) |
- |
W |
12–40 (20) |
- |
Ratio of right spicule to body length |
1:3-1:4 (1:4) |
- |
Ratio of left spicule to body length |
1:2–1:3 (1:3) |
- |
Distance of anus from tail tip |
100– 225 (149) |
351–468 (411) |
Tail process L |
70–74 (72) |
49–126 (81) |
Tail process W |
23–39 (31) |
18–72 (42) |
Pre–cloacal Papillae No. |
28–32 (14–16 pairs) |
– |
Paracloacal papillae No. |
1pair sessile+1pair foliate papilla |
– |
Post–cloacnal papillae No. |
12–14 (6–7 pairs) |
– |
Spines at tail tip L |
7–10 (8) |
7–14(10) |
Ratio of ventricular appendix Vs body length |
1:6–1:7 (1:7) |
1:6–1:8 (1:6) |
Ratio of intestinal ceca Vs ventricular appendix |
1:2–1:4 (1:3) |
1:1–1:4 (1:2) |
Ratio of width of ventricular appendix Vs length of ventricular appendix |
1:8–1:11 (1:107) |
1:8–1:10 (1:9) |
Foot note: Range (Mean) |
Table 1 Morphometric measurements of adult worms of Indospinezia multispinatum gen. et sp. n
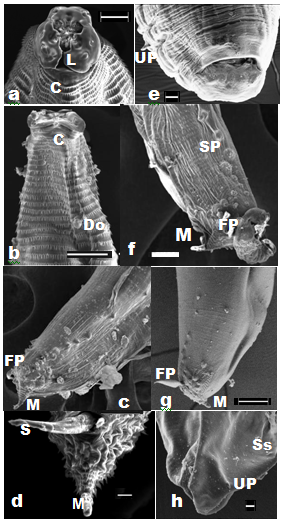
Pattern of spination
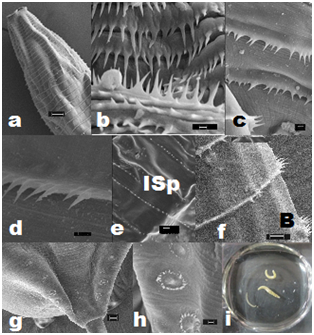
Sharp elongated single-toothed unidentate spines, of varied sizes, were present, and their arrangement differed in distinct horizontal rows on the cuticular envelope, covering the body surface post-base of the head (Figures 2a,2b&4b-4f). The spines were devoid of any specialized anatomical feature in the region of oesophageo-intestinal junction. Irregularly alternate placement of longer and smaller-sized spines in each row, particularly between the 63rd and the 104th, were noticed. In addition, irregularly spaced clusters of two to four spines, on a muscular base, with single-toothed unidentate elongated spines and a few smaller spines, in between, were also recorded Figure 3d. The peculiar composition of paired spines and irregularly-spaced single spines, in between, in the same row, were observed in the hinder part of body before the anal region. Incomplete rows of minute spines could be seen intermittently on the body. The knob-like roundish bosses with cluster of unequal spines occurred in mid-body rows between cuticular transverse striations on lateral side of body, on the body of adult worms.
The total number of horizontal rows of spines was 174-225 (200±8) in males and 201-305 (298±10) in females, on whole body of worms, as well as their larvae under study. The spines were minimum in number, shorter as well as in congested rows in the immediate post-cephalic region, varying from 104-128 (116±12) in each row in female worms, to 120-140 (130±10) in males. The minute rows of spines bordering the rim of cloacal aperture with sparsely distributed longer spines in the paracloacal region were distinctly visible. The measurements of spines on various parts of the body of the worm are given in Table 2.
Males |
|
Females |
|
|---|---|---|---|
No. of rows |
Measurements of spines (mm) |
No. of rows |
Measurements of spines (mm) |
1–15th row L |
11–15 |
1–15th row L |
15–25 |
W |
2–4 |
W |
4.3–8 |
16–68th row L |
16–33.5 |
16–66th row L |
28–32.4 |
W |
3.9–11 |
W |
7–7.8 |
69–137th row L |
11.7–27.3 |
67–83rd row L |
36–39 |
W |
3.9–11.7 |
W |
9–9.8 |
138–143th row L |
3.9–11.7 |
84–199throw L |
19–19.5 |
W |
0.7–3.9 |
W |
3.9–5.1 |
144–192nd row L |
5.8–7.8 |
200–257th row L |
11–12 |
W |
1.2–1.6 |
W |
3.9–4.7 |
193–225th row |
Minute spines |
258–288th row L |
3.9–6.2 |
W |
1.9–3.1 |
||
|
|
288–305th row |
Minute spines |
Table 2 Measurements of spines on body of adult worms of Indospinezia multispinatum gen. et sp. N
Spination pattern on male
Maximum number of spines, 278-288 occurred in 29th-49th row at the level of ventriculus extending upto 2/3rd length of ventricular appendix. The distance between adjacent rows of spines, 121-172 was the maximum in the mid-body region between 99-119th rows from the end. Initially the distance between two rows of spines from 1st to 6th rows was 8-16; at the level of ventriculus, 12-117; between 120-177th rows in posterior half of body, 117-152, and between 172-192nd rows of spines near cloacal region, 47-62. The length of spines started declining from 68th row onwards, and only minute spines were visible between the 193rd and 225th rows adjacent to the posterior extremity.
Maximum number of spines, 394-402 was recorded at 113th -114th row. The distance between adjacent rows of spines, 94-133, was maximum at about anterior one-fourth of the length of body. The distance was, 12-23 between 1st-15th rows of spines at the anterior end; 27-49 between 16-31st rows; 66-117 between 99-150th rows; 59-125 between 151st-209th rows, and 39-59 between 210th-288th rows, and very minute spines were observed between 289th-305th rows. The length of spines started declining from 83rd row.
Male
Male worms were large. Two long, alate sub-equal spicules (Figure 5d), 2.68-2.85% (2.74±0.05%) - 3.19-3.64% (6.38±0.15%) of the length of body of worms. The caudal papillae comprised 6-7 pairs of post-cloacal and 14-16 pairs (28-32 no.) of pre-cloacal (Figures 2b,2d,2e&5d). A paired foliate para-cloacal papillae were present near anus along with a sessile pair lateral to cloacal aperture, and an unpaired phasmid at the base of the tail, that ended in a short terminal mucronate (Figures 2d,2f,2g). The pre-cloacal papillae comprised one pair of dorso-lateral foliate papillae Figure 2g located a short distance before anus. Two pairs of larger papillae with specialized muscular arrangement were followed by a series of 11pairs of ‘sunflower’ pre-cloacal papillae (Figure 2c,2g&3c) after which another set of two pairs of larger papillae with specialized muscular arrangement followed. A basal pair of parallel running elongated muscular flaps, arranged longitudinal to the body plane, overlapped the ring of spines at the base of these papillae. Two unpaired additional papillae were also observed in the vicinity of phasmid.
Female
Female worms measure, 130-1805(1593+121)x49-243 (114+34). Vulva pre-equatorial varied from 33-43% of body length of females. The typical caudal papillae were on tail, one pair of these were pedunculate, ungulate (Figure 2h&4g,4h), while the other pair was sessile, and a short terminal mucronate was present at the tail end with a phasmid at the basal rim of cloaca (Figure 2e).
Differential diagnosis
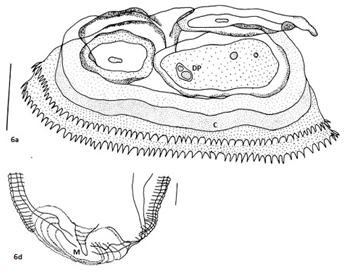
Larvae with cephalic hooks or spines; multinucleate oesophageal glands (Figure 3e) (Figure 3f); cuticle with transverse annulations bearing transverse rows of single-toothed unidentate spines of variable shapes and patterns of spination; oesophagus comprised of an anterior muscular and posterior glandular part; two pairs of cervical sacs; eggs with polar filaments at one end. Three cephalic lips; ventricular appendix, and the ‘cactus’ tail, similar to the one observed in genus Goezia15,33 distinguished these worms from spirurines. In addition, a post-labial, elevated double-ridged collarette at the base of head, was the typical character of Physalopteridae, while frontal sclerotized lid-plate to cover triangular oral cavity apically Figure 6a, was a unique feature that differentiated these from spirurines, raphidascaridoid as well as the worms of Physalopteridae. The unique morphologically peculiar pre-cloacal papillae and pattern of spination on body surface of the newer worms differentiated it from the gnathostomatoid worms. In addition, the number of caudal papillae (23-24pairs pre-cloacal vs 7-9pairs), post-cloacal double papilla not observed (one pair in G. Bangladeshi), two pairs of pre-cloacal double papillae and foliate papillae each, were the differential features of the newer worms.
18S rRNA, ITS-1 and ITS-2 gene analyses
The 18S rRNA gene sequence analysis revealed 0.004 average K2P distance of individuals within I. Multispinatum gen. et sp. n. (Nematoda: Anisakidae) as compared with an average distance of 0.037 of the genus Goezia and 0.043 of Gnathostoma. The two genera with which the present worms showed critical morphological similarities. The average K2P distance was 0.004 vis-à-vis 0.008-0.067 between individuals within I. multispinatum gen. et sp. n. based on ITS1 gene sequence, and individuals of other closer genera. Simultaneously, it was 0.002 vis-à-vis 0.010-0.041 between individuals within I. Multispinatum gen. et sp. n. based on ITS2 gene sequence, and individuals of other closer genera. Noticeably, the 863bp long 18S region possessed 838(97.1%) conserved domains and 14 parsimony informative sites within the same organism, in which the while 545bp long ITS-1 region possessed 522(95.6%) conserved domains and 14 parsimony informative sites, and 465bp long ITS-2 region possessed 459(98.7%) conserved domains and 30 parsimony informative sites.
coi gene analyses
The average K2P distances based on Mit. coi analyses between I. Multispinatum gen. et sp. n. and members of the genera Contracaecum, Hysterothylacium, Gnathostoma and Parascaris were 0.018, 0.003, 0.025 and 0.029, respectively. However, the value between I. Multispinatum gen. et sp. n. and Raphidascaris and Anisakis was 0.043. The 655bp long Mit. coi region possessed 622 conserved domains (94.96%) and 17 parsimony informative sites.
Analysis of GC content
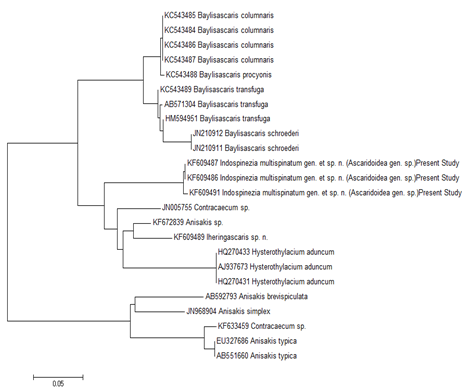
The GC content variations in 18S rDNA, Mit. coi, ITS1 and ITS2 genes as shown in Table 3, were consistent within individual genes between all the different individuals on which the investigations were conducted. The overall GC content of the new genus I. Multispinatum gen. et sp. n., based on 18S rDNA, Mit. coi, ITS1 and ITS2 gene compositions, ranged from 34.1-54.4%. Therefore, the barcode structure, including detailed analysis of phylogeny of worms, based on information on 18S rDNA (Figure 6), ITS1 Figure 7, ITS2 (Figure 8), and Mit. coi (Figure 9) genes, were studied. The inference was evaluated and interpreted to support the contention of inclusion of the collected worms under the newly proposed genus, Indospinezia gen. n. as a new species, I. Multispinatum gen. et sp. n.
Name of the worm and accession No. |
Total |
1st base |
2nd base |
3rd base |
ITS1 |
||||
KF609486 Ascaridoidea gen sp (A) |
50.5 |
55.4 |
45 |
50.8 |
KF609487 Ascaridoidea gen sp (A) |
51.8 |
57.9 |
47.1 |
50.3 |
KF609491 Ascaridoidea gen sp (L) |
51.7 |
55.9 |
45.9 |
52.8 |
ITS2 |
||||
KF609493 Ascaridoidea gen sp (A) |
54.3 |
59.3 |
52.6 |
50.7 |
KF609494 Ascaridoidea gen sp (A) |
54.4 |
60.3 |
52.9 |
50 |
KF609490Ascaridoidea gen sp (L) |
54.2 |
60.3 |
51.9 |
50.3 |
18S rDNA |
||||
KF609495 Ascaridoidea gen sp (A) |
48.3 |
44.4 |
49.3 |
51.2 |
KF609496 Ascaridoidea gen sp (A) |
48.9 |
44.3 |
48.7 |
52.5 |
KF609492 Ascaridoidea gen sp (L) |
48.6 |
44.3 |
49.3 |
52.2 |
GQ265676 to GQ265684 Spiruroidea gen sp (A) |
48.7 |
44.3 |
49.4 |
52.1 |
Mit. coi |
||||
FJ172981 Spiruroidea gen sp (A) |
34.1 |
31.1 |
34.1 |
37 |
Table 3 GC content percentage at 1st, 2nd & 3rd base positions in different gene sequences of Indospinezia multispinatum gen et sp. N

The outbreak of worms from marine fish with identical genetic characterization to those of the worms from freshwater fish in the Gangetic riverine ecosystems were reported in India recently.36 The systematics of Ascaridoids was reorganized to comprise 5 families and 8 subfamilies,19 based on the structure of secretory-excretory system, in combination with the established concepts.37 Later, several workers38,39 used barcode structures to propose phylogenetic hypotheses for some ascaridoids, and the phylogenetic trees were developed with the help of ribosomal DNA sequences. These trees supported the classification of Ascaridoidea19 proposed earlier, though the detailed illustrations later inferred13 that rDNA based genetic characterization was heavily biased towards a single locus, and therefore, certain queries remained unanswered.
The genus Goezia40 was conclusively placed among Raphidascarididae with 99% bootstrap support vide analysis of the transversion-weighted cox2 gene tree generated.5 The primary significance of life-cycle patterns, morphology of the oesophagus and female reproductive organs in nematode taxonomy was highlighted,9,19 in addition to genetic characterization. In this light, the site specificity of the adult worms and larvae of the new genus, post-migration of developmental stages assumed greater significance. The morphology and life-cycles were also heavily relied upon by several earlier prominent workers.16,19,36‒40 to raise several classifications to date. The experimental data on the life cycle stages of the newer worms was, therefore, generated, and it has been communicated as a separate manuscript along with of these, genus Lappetascaris was known to be the only genus under subfamily Lappetascaridinae whose status was altered from that of a family to tribe.42 This genus9 was recently transferred19 as a synonym of Hysterothylacium (Ward and Magath). Although the ‘larval tooth’ was not reported15 in second stage larvae of G. bangladeshi, whose juveniles were only found in their study in the gut of fish host, Tenualosa ilisha, yet it was attempted to draw parallel between its life cycle with H. aduncum (Rudolphi),11 that bore larval tooth in the larval stage, on the basis of advanced stage larvae (4th or young 5th stage), i.e. the juveniles of G. Bangladeshi. Therefore, the presumption on the life cycle stages of G. Bangladeshi deduced in the earlier stud15 were contradictory to the experimentally produced larvae of G. Ascaroides 43‒46 with boring tooth in second stage larvae.47 The affinity of the newer worms with spirurin nematodes, and therefore, Gnathostomatoidea (Railliet),48 was apparent due to 3 main reasons- (i) their larval forms possessing cephalic hook,49,50 (ii) oesophageal glands being multinucleate and (iii) their characteristic of moulting larvae within the eggs. The literature on the role of fish as a definitive host to gnathostomatoid nematodes has been scarce.49 Gnathostomatoidea was the only one out of the four super families of the suborder Spirurina, to which the newer worms showed closer affinity, whose members were parasitic as adults in freshwater fish,43 but liver as the site of infection was specific in the present worms.
The infections by newer worms neither occurred in the alimentary tract nor any nodules in the gut wall were formed to contain developing larvae, while these were the consistent features of worms in the earlier study.15 Though the experimental development through a copepod intermediate host has been propagated,15 but contrary to this, the role of the same fish host as an intermediate as well as the definitive host has been reported in the present study in natural infections. These findings derived support from the earlier reports9,37 that the occurrence of obligate copepod intermediate host was not mandatory. This eventuality simultaneously explained autogenicity of infections in the riverine ecosystem wherein host-specific infections in X. Cancila were encountered. The need for extensive investigations to reveal intricacies of the life cycles of parasitic adult gnathostomatoid worms in fish to point out the level of their significance in establishing systematics of worms, has been expressed already.49 The concept of precocity was propounded in G. Bangladeshi as well,15 and it was claimed to be similar to H. Aduncum in earlier studies51 on ascaridoids, and in several species of Gnathostoma.19,50‒52 It was, however, categorically emphasized53 that the third stage larvae increase enormously, at times more than three times50 in size, to become AdvL3, where fourth-stage larvae were absent in the life cycle.50 The evidence of similar significant increase in the size of third stage larvae has been recorded in the life cycle of the present worms that has been communicated separately along with this manuscript.16 Contrary to this, when fish spirurines employed a paratenic host, the third-stage larvae usually did not grow considerably. It was thus suggested that such unusual preparatory growth in AdvL3 was compensated for the absence of the fourth larval stage. Therefore, in this crucial aspect of absence of AdvL4 and AdvL515 in life cycle, the worms of the new genus differed substantially from fish spirurines and anisakid worms, besides several other strikingly different morphological features, as illustrated in the foregoing text. The sexually mature specimens of G. Bangladeshi were considered to have represented precocity, because of their recovery from the cavities of the stomach wall of T. Ilisha15 to merit the fish being considered as an intermediate or transport host, and not a definitive host. The specimens of the new genus collected from the hepatic cavity of fish quite frequently included gravid females possessing vulva, along with sexually mature males that possessed fully developed ‘cactus’ tail with ‘sunflower’ genital papillae and spicules. This also confirmed the ‘definitive host’ status of the fish. This is thus amply clear that AdvL3 of the worms of the new genus moulted to the adult stage directly in definitive host, as also emphasized.15,36 This would also mean that the life cycle of the newer worms comprised total four stages - three larval and one adult forms, and there was no fourth or fifth stage larva in I. Multispinatum gen et sp. n.The events parallel to the occurrence of life cycle stages of the newer worms in practically every body organ of the host fish were encountered in some of the species of genus Goezia. The latter have been known to detach from the site of infection in the wall of gut, and wander around, before returning to the original site of infection in the alimentary canal,48 as was the progression of developmental stages of the new genus to migrate to the site of infection, i.e. liver of fish. The former fed on host tissue and the predigested food inside alimentary canal, though the latter survived substantially on hepatic tissues, and fed on a variety of muscular and other visceral tissues, but alimentary canal was not the site of infection of the newer worms.
The remarkably differentiating features of the closer worms from genus Goezia were a sclerotized lid-plate atop sclerotized stoma orally, oesophagus with an anterior muscular and a posterior glandular part, 2 pairs of cervical sacs, and the structurally different ‘sunflower’ caudal papillae, that comprised specialized trilaminate muscular arrangement with spiny rings one over the other, around these papillae, with a pedunculated papilla atop (Figures 2f,2g,3c), and a basal pair of parallel running elongated muscular flaps, one longer than the other, arranged longitudinally, that overlapped the above trilaminate structure. The newer worms also differed in smaller ventriculus as percent of body length (1.63-2.85% Vs 10.61-15.92%), ventricular appendix 7-11times as long as wide, the excretory pores being at a farther distance (21-23rd rings Vs 7th ring), and the newer worms being parasitic in tissues and spaces of the freshwater fish.
The morphological differentiation of the newer worms with all the valid genera under Anisakidae was distinctly established. These differed from Pulchrascaris, Terranova, Pseudoterranova, Contracaecum, Phocascaris, Amphibiogoezia, Heterotyphlum, Paraheterotyphlum, Thynnascaris, Iheringascaris, Ichthyascaris, Raphidascaroides, Alibagascaris, Maricostula, Mehdiascaris, Ortoanisakis and Mawsonascaris in the presence of sclerotized lid-plate atop sclerotized stoma orally, an amphid on the dorsal lip, 2 pairs of cervical sacs, a bitubular ventricular appendage, and the ‘sunflower’ pre-cloacal papillae. The worms of the newer genus resembled closely to genus Goezia40 in the presence of mouth with flattened lips extending outwards, separated from the body, ventriculus with long, double appendix, an intestinal caecum, and “sunflower” caudal papillae, but differed from it in the peculiar pattern of spination on body, typical pattern of distribution as well as number of pre- and post-cloacal papillae in male worms. The worms of the newer genus resembled closely to Pulchrascaris (Vicente and Dos Santos) in similarity of habitat within liver of definitive host, but differed too from it in possessing reduced lips, non-alate spicules, in absence of cuticular plates immediately posterior to cloaca; Terranova in possessing a ventricular appendix and in absence of median transverse cuticular plates posterior to cloaca; Pseudoterranova in possessing a post-labial, elevated double-ridged collarette at the base of head, the unique morphologically peculiar pre-cloacal papillae, pattern of spination on body surface, absence of denticulate caudal plates and the life cycle not passing through an intermediate crustacean host. The worms of the newer genus were also similar to Contracaecum in possessing a folded cuticular collar present at distal margin of cephalic region, but differed from it in having higher ratio of caecum length to appendix length, being upto 1:4(Vs 1:2), absence of interlabia, and from Phocascaris (Synonym to Contracaecum) in absence of interlabial knobs, excretory pore situated immediately posterior to ventral interlabial knob, inner margin of each lip with median indentations, though I. Multispinatum gen. n. et sp. n. resembled closely with Phocascaris in terms of cuticle behind lips formed collar, and 80-100 cloacal papillae as innumerable pre-cloacal papillae have been reported in the former worms. The newer worms, however also differed in being parasitic in the intestine of X. cancila than from the stomach of hooded seal, Crystophora Cristata in which worms of were found; from Amphibiogoezia in possessing spinulated annules prevalent all over the length of body of worms, absence of gubernaculum, and precloacal sucker, and parasitization restricted to fish as intermediate as well as definitive host; from Heterotyphlum in absence of dentigerous ridges on lips, presence of a bitubular ventricular appendage, spicules subequal, and the spines on tail; from Sprentascaris in presence of interlabia, a bitubular ventricular appendage, excretory pore open behind nerve ring, spicules sub-equal, and the spines on tail and Paraheterotyphlum in possessing lips that were not rectangular, and a bitubular ventricular appendage; Thynnascaris in possessing a in the ratio of caecum length to appendix length being upto 1:4(Vs 1:1) and absence of interlabia. It was, however, significant that this genus was closer to Indospinezia gen.n. in the presence of well developed gonads in the posterior body of Thynnascaris sp. (Type IV) larva, as reported;54 Iheringascaris in possessing a typical pattern of spination on cuticle on the whole body surface, in absence of interlabia and unequal spicules and being parasitic in freshwater fish; Ichthyascaris in possessing an intestinal caecum, non-alate spicules, and in absence of flanges posterior to sub ventral lips; from Raphidascaroides in absence of dentigerous ridges on lips, presence of a bitubular ventricular appendix; Alibagascaris in possessing lips without dentigerous ridge, oesophagus with an anterior muscular and a posterior glandular part, unequal spicules, in absence of gubernaculums, and being parasitic in freshwater fish; Sprentascaris in possessing unequal spicules; Maricostula in possessing lips with interlabia, and elongate intestinal caecum, pyriform ventriculus, long sac-like ventricular appendix, as well as greater number of cloacal papillae in male worms; Mehdiascaris in possessing a vulva in absence of lappets at the proximal margins of lip and being parasitic in freshwater fish; Ortoanisakis in possessing interlabia, absence of both the ventricular appendix and intestinal caecum and lesser number of caudal papillae. However, the genus Ortoanisakis was synonymized with Paranisakis.42 The new genus was comparable with Mawsonascaris in possessing greater number of cloacal papillae, in absence of dentigerous ridges and gubernaculum and being parasitic in freshwater fish. The newer worms further differed from Raphidascaris because of the presence of papillae between the rows of spines on body at the level of 15th, 18th and 20th row from anterior end, a pair of deirids and the spines on tail; from Hysterothylacium in possessing in possessing a typical pattern of spination on cuticle on the whole body surface, in absence of interlabia, the specialized pattern of distribution of caudal papillae along with unique ‘sunflower’ papillae and spines on the tail. The affinity of Goezia with Raphidascarididae was established5 by the application of combined evidence tree, but subfamily Goeziinae19,55 was recognized to include genus Goezia and the Tribe Raphidascarididae under subfamily Raphidascaridinae56 to include genus Raphidascaris (Railliet and Henry). On the basis of the genetic heterogeneity, the worms of the new genus would stand out exclusive in the present study, as depicted by separate clades of ITS1 Figure 22, ITS2 Figure 8 gene sequences of the present worms from those of H. Aduncum (Raphidascarididae) and Anisakinae. In light of this, it is proposed to split family Goeziinae into two new tribes, to accommodate, firstly the new genus, Indospinezia gen. n. under the newly proposed tribe, Indospineziinea Tribe n., and secondly, genus Goezia be included under a newly raised Tribe Goeziinea Tribe n.
Gene sequence analyses
As is evident, the 12 sequences obtained from 18S rDNA gene analyses of the representative specimens of mature worms andAdvL3 of the new genus, showed that these fell on a separate branch in the NJ tree Figure 6 forming a separate clade, and hence were different from all other organisms under study Table 4. The monophyletic characterization of the new genus was thus evident. It is pertinent to mention that the sequences obtained from ITS1 Figure 7 and ITS2 Figure 8 gene analyses of the representative specimens of two mature worms and one AdvL3 fell in separate clades that were distinct from other ascaridoids, whose sequences were available at NCBI. The sequence obtained from Mit. coi gene analysis of the representative specimen of mature worm also exhibited a separate branch FJ172981, Figure 9, that was distinct from Gn. binucleatum (AB037131) and Gn. spinigerum (AB037132) on one hand, and Hysterothylacium (FJ907319) and Contracaecum (FJ907321) on the other. It did not show significant similarity with other phylogenetically closer genera as revealed by the bootstrap values i.e 1000 replicates Figure 9, and also by morphometric studies.
The mystery of evolutionary lineages could be revealed by the investigations on mtDNA.57 The nuclear ex-mitochondrial sequences have recently been suggested to be a powerful tool in various aspects of evolutionary biology, as the studies on nuclear integrations have suggested that the relative rates of evolution of mtDNA and single-copy nuclear DNA (scnDNA) have major significance for their use as molecular markers in evolutionary studies. The sequences generated from the analyses conducted during the study did not comprise any insertion or deletion, nor were the stop codons detected. Therefore, the findings on Mit. coi gene analysis corroborated the assertions published earlier57 that since the sequences were devoid of stop codons, none of the pseudogenes or NUMTs (nuclear DNA sequences originating from mitochondrial sequences) were obtained to attain desired gene sequencing.
Genbank accession no. |
Gene |
Taxon |
Reference |
KF609487 |
ITS1 |
Indospinezia multispinatum gen. et sp. n. |
Present study |
KF609491 |
ITS1 |
Indospinezia multispinatum gen. et sp. n. |
Present study |
KF609486 |
ITS1 |
Indospinezia multispinatum gen. et sp. n. |
Present study |
KC543488 |
ITS1 |
Baylisascaris procyonis |
|
GU295975 |
ITS1 |
Anisakis paggiae |
|
AB592793 |
ITS1 |
Anisakis brevispiculata |
|
JN968904 |
ITS1 |
Anisakis simplex |
|
KF633459 |
ITS1 |
Contracaecum sp. |
|
EU327686 |
ITS1 |
Anisakis typica |
|
KF672840 |
ITS2 |
Anisakis sp |
|
KF633463 |
ITS2 |
Iheringascaris |
|
KF609490 |
ITS2 |
Indospinezia multispinatum* gen. et sp. n. |
Present study |
KF609494 |
ITS2 |
Indospinezia multispinatum* gen. et sp. n. |
Present study |
KF609493 |
ITS2 |
Indospinezia multispinatum* gen. et sp. n. |
Present study |
KF633462 |
ITS2 |
Contracaecum sp.n. |
|
AB428418 |
ITS2 |
Gnathostoma binucleatum |
|
FJ524380 |
ITS2 |
Gnathostoma turgidum |
|
FJ524381 |
ITS2 |
Gnathostoma turgidum |
|
EU930822 |
ITS2 |
Gnathostoma turgidum |
|
AB551660 |
ITS1, ITS2, |
Anisakis typical |
|
HQ270433 |
ITS1, ITS2, |
Hysterothylacium aduncum |
|
AJ937673 |
ITS1, ITS2, |
Hysterothylacium aduncum |
|
HQ270431 |
ITS1, ITS2, |
Hysterothylacium aduncum |
|
JN210912 |
ITS1, ITS2, |
Baylisascaris schroederi |
|
JN210911 |
ITS1, ITS2, |
Baylisascaris schroederi |
|
AB571304 |
ITS1, ITS2, |
Baylisascaris transfuga |
|
HM594951 |
ITS1, ITS2, |
Baylisascaris transfuga |
|
KC543489 |
ITS1, ITS2, |
Baylisascaris transfuga |
|
KC543487 |
ITS1, ITS2, |
Baylisascaris columnaris |
|
KC543486 |
ITS1, ITS2, |
Baylisascaris columnaris |
|
KC543485 |
ITS1, ITS2, |
Baylisascaris columnaris |
|
KC543484 |
ITS1, ITS2, |
Baylisascaris columnaris |
|
JN005755 |
ITS1, ITS2,18S rDNA, |
Contracaecum sp |
|
U94377 |
18S rDNA |
Iheringascaris inquires |
|
GQ265670 |
18S rDNA |
Iheringascaris inquires |
|
DQ503460 |
18S rDNA |
Raphidascaris acus |
|
JF718550 |
18S rDNA |
Hysterothylacium deardorffoverstreetorum |
|
U94376 |
18S rDNA |
Hysterothylacium reliquens |
|
U94374 |
18S rDNA |
Hysterothylacium fortalezae |
|
U94375 |
18S rDNA |
Hysterothylacium pelagicum |
|
U94380 |
18S rDNA |
Pseudoterranova decipiens |
|
EF180082 |
18S rDNA |
Anisakis pegreffi |
|
U94365 |
18S rDNA |
Anisakis sp |
|
EF180074 |
18S rDNA |
Porrocaecum streperae |
|
AF036608 |
18S rDNA |
Toxocara canis |
|
JN256975 |
18S rDNA |
Toxocara cati |
|
EF180078 |
18S rDNA |
Toxocara vitulorum |
|
EF180059 |
18S rDNA |
Toxocara cati |
|
JN256982 |
18S rDNA |
Toxascaris leonine |
|
*Ascaridoidea, and # Spiruroideaat NCBI |
18S rDNA |
Porrocaecum angusticolle |
Honisch & Krone (2008) |
GQ265684 |
18S rDNA |
Indospinezia multispinatum# gen. et sp. n. |
Present study |
GQ265683 |
18S rDNA |
Indospinezia multispinatum# gen. et sp. n. |
Present study |
GQ265682 |
18S rDNA |
Indospinezia multispinatum# gen. et sp. n. |
Present study |
GQ265681 |
18S rDNA |
Indospinezia multispinatum# gen. et sp. n. |
Present study |
GQ265680 |
18S rDNA |
Indospinezia multispinatum# gen. et sp. n. |
Present study |
GQ265679 |
18S rDNA |
Indospinezia multispinatum# gen. et sp. n. |
Present study |
GQ265678 |
18S rDNA |
Indospinezia multispinatum# gen. et sp. n. |
Present study |
GQ265677 |
18S rDNA |
Indospinezia multispinatum# gen. et sp. n. |
Present study |
GQ265676 |
18S rDNA |
Indospinezia multispinatum# gen. et sp. n. |
Present study |
GQ265671 |
18S rDNA |
Rostellascaris sp |
|
U94366 |
18S rDNA |
Ascaris lumbricoides |
|
JN256991 |
18S rDNA |
Baylisascaris ailuri |
|
DQ442661 |
18S rDNA |
Terranova scoliodontis |
|
U94368 |
18S rDNA |
Baylisascaris procyonis |
|
JN256992 |
18S rDNA |
Baylisascaris schroederi |
|
JN256989 |
18S rDNA |
Baylisascaris transfuga |
|
U94369 |
18S rDNA |
Baylisascaris transfuga |
|
JN256984 |
18S rDNA |
Toxascaris leonine |
|
JN256977 |
18S rDNA |
Toxocara canis |
|
U94378 |
18S rDNA |
Parascaris equorum |
|
U94383 |
18S rDNA |
Toxascaris leonine |
|
JF803924 |
18S rDNA |
Goezia spinulosa |
|
U94372 |
18S rDNA |
Goezia pelagia |
|
U94379 |
18S rDNA |
Porrocaecum depressum |
|
KF633454 |
18S rDNA |
Anisakidae |
|
KF633453 |
18S rDNA |
Anisakidae |
|
KF633456 |
18S rDNA |
Anisakidae |
|
KF672838 |
18S rDNA |
Anisakis larva |
|
KF609492 |
18S rDNA |
Indospinezia multispinatum* gen. et sp. n. |
Present study |
KF609496 |
18S rDNA |
Indospinezia multispinatum* gen. et sp. n. |
Present study |
KF609495 |
18S rDNA |
Indospinezia multispinatum* gen. et sp. n. |
Present study |
GQ132133 |
COI |
Anisakis simplex |
|
GQ132129 |
COI |
Anisakis simplex |
|
GQ132125 |
COI |
Anisakis simplex |
|
DQ489705 |
COI |
Anisakis simplex |
|
GQ132128 |
COI |
Anisakis simplex |
|
GQ132124 |
COI |
Anisakis simplex |
|
AB551552 |
COI |
Gnathostoma spinigerum |
|
AB576759 |
COI |
Pseudoterranova azarasi |
|
AB576758 |
COI |
Pseudoterranova azarasi |
|
HM594948 |
COI |
Baylisascaris transfuga |
|
KF536871 |
COI |
Ascaris lumbricoides |
|
KF536870 |
COI |
Ascaris lumbricoides |
|
KF536863 |
COI |
Ascaris lumbricoides |
|
KF536862 |
COI |
Ascaris lumbricoides |
|
KF536861 |
COI |
Ascaris lumbricoides |
|
KF536860 |
COI |
Ascaris lumbricoides |
|
AF179917 |
COI |
Iheringascaris inquires |
|
HM147278 |
COI |
Pseudoterranova decipiens |
|
FJ172981 |
COI |
Indospinezia multispinatum gen. et sp. n. |
Present study |
Table 4 Representative groups of ITS-1, ITS-2, 18S rDNA and cytochrome c oxidase (coi) gene sequence selected for the study
The affinity of Spiruromorpha with Ascaridomorpha, in terms of the conception of earlier parasitologists17 was distinctly illustrated in an extensive Mit. coi genome-based framework Figure 9, wherein a specific clade emerged encompassing the newer worms, Gn.spinigerum, G. Binucleatum, Contracaecum and H. Aduncum. This was further supported by a clade that comprised as many as 4 genera of Ascaridomorpha (Anisakis, Pseudoterranova, Baylisascaris and Ascaris) with Spiruromorpha represented by Gnathostoma Figure 9. The newer genus Indospinezia gen. n. (11 sequences) became paraphyletic with other genera of Ascaridomorpha, viz. Goezia represented by 2 species; Rostellascaris by 1 species; Hysterothylacium by 4 species; Raphidascaris by 1 species; Iheringascaris by 2 species; Toxascaris by 1 species; Toxocara by 3 species; Pseudoterranova by 1 species; Anisakis by 3 species; Ascaris by 1 species; Porrocaecum by 1 species; Parascaris by 1 species; Baylisascaris by 4 species; and Terranova by 1 species with Contracaecum sp. having emerged as an out group Figure 6.
Analysis of the GC content
The variations in GC content in 18S rDNA and Mit. coi genes showed higher content at third position and minimum at first position, while the reverse was observed in for ITS2 gene Table 5. However, the divergence in GC content was the highest at first position in ITS1 and ITS2 genes. The association of the augmented genetic divergence with level of radiation that occurred in parasites, as the parasitic life cycle progressed, was advocated in earlier studies.58 The mitochondrial molecular divergence of G+C content, noticeably different from ITS1, ITS2 and 18S rDNA genes, as recorded in the present investigations, were linked to the extraordinary level of radiation that occurred in parasitic organisms.59 The inference, therefore, brought closer the possibility of divergence in the worms of the new genus to the divergent characteristics of Anisakidae, fish spirurines, particularly gnathostomatoid and physalopteroid worms on the basis of their morphological and genetic characterization. It has already been put forth60 that gnathostomatoid worms represented a very early branch of the Spirurida. In those terms, the newly raised tribe proposedly acted as a connecting link before the worms of Ascaridoidea evolved. Therefore, on the basis of the aforementioned significant points of morphological and genetic characterization, it is proposed to accommodate the worms of the new genus as, I. Multispinatum gen et sp. n. (Nematoda: Spiruroidea) under a new subfamily, Indospineziinae subfam. n. This study thus vindicated the opinion expressed recently61 on the presentation of integrated molecular data to supplement analysis of robust taxonomic assessment with morphological details of ascaridoid worms.
The affinity of the raphidascaridoid nematodes, Indospinezia multispinatum gen. et sp.n., belonging to the newly proposed subfamily Indospineziinae subfam., with anisakid worms, particularly G. Bangladeshi, and spirurins, particularly Gnathostomatoid worms, was intriguing. The typical anisakid ventriculus at the oesophageo-intestinal junction, ‘sunflower’ caudal papillae on ‘cactus’ tail similar to those reported in the former species, and striking spirurin features, viz. cephalic hook in larvae, oesophageal glands being multinucleate and moulting larvae within the eggs were the noticeable features. However, the peculiar differentiating distribution pattern of precloacal papillae, liver being the site of infection by larvae as well as adult worms, the presence of sclerotized lid-plate atop sclerotized stoma orally, an elevated post-labial double-ridged collarette at the base of the head, demarcating from rest of the body, an amphid on the dorsal lip, intestinal caecum, oesophagus comprising an anterio-muscular and posterior-glandular part, and 2 pairs of cervical sacs were the differentiating characteristics of the newer worms from all the closer genera of Raphidascarididae. AdvL3 of the worms of the new genus moulted to the adult stage directly in definitive host, as also emphasized.49,50 This would also mean that the life cycle of the newer worms comprised total four stages - three larval and one adult forms in I. Multispinatum gen et sp. n. Therefore, morphological and genetic characterization based on 18S rDNA, Mit. ITS1 and ITS2 gene analysis, the newer genus is proposed to be accommodated under the newly raised tribe Indospineziinea tribe n. under the family Raphidascarididae. The subfamily Raphidascaridinae42 that was also upheld as valid10 is reinstated. The findings17 based on SSU rRNA-based results warranted a new delimitation of genera, to fulfill the requirement of a broad taxonomic revision, supplemented by additional morphological and molecular characters that have been addressed to in the current investigation. The findings of the present investigation derived strong support from the assertion of authors13 for the conclusion of I. Multispinatum gen et sp. n. being ancestor for Spiruromorpha and Ascaridomorpha, as it was emphasized13 that the worms possessing monoxenous life cycle are ancestral to the group.
The nucleotide sequence data of mature worms of I. Multispinatum gen et sp. n. (Nematoda: Spiruroidea) reported in this study are available in Genbank database under the accession numbers, KF609486, KF609487, KF609493, KF609494, KF609495, KF609496, GQ265676, GQ265677, GQ265678, GQ265679, GQ265680, GQ265681, GQ265682, GQ265683, GQ265684 and FJ172981, and of AdvL3 under the accession numbers, KF609490, KF609491, and KF609492.
The authors appreciate facilities in the Department of Zoology, Allahabad Central University and Department of Zoology, Nehru Gram Bharati University at Allahabad, U.P, India to carry out the work. The assistance to procure scanning electron photomicrographs by USIF at Aligarh Muslim University is greatly appreciated. The assistance of periodical fish catches from the riverine ecosystems at Allahabad, received from the fisherman Sri Pattu and his colleagues is acknowledged.
Author declares that there is no conflict of interest.

©2017 Jaiswal, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.