International Journal of
eISSN: 2573-2838


Mini Review Volume 4 Issue 5
Department of Agrifood and Biology, Institute of Science of Foods Productions of National Research Council (ISPA-CNR), Italy
Correspondence: Palmiro Poltronieri, Department of Agrifood and Biology, Institute of Science of Foods Productions of National Research Council (ISPA-CNR), via Monteroni km 7, 73100 Lecce, Italy, Tel +39 0832 422609
Received: October 03, 2018 | Published: October 30, 2018
Citation: Poltronieri P. Polyhydroxyalkanoate production in biofermentor monitored through biosensor application. Int J Biosen Bioelectron. 2018;4(5):23510.15406/ijbsbe.2018.04.00133
Background
Polyhydroxyalkanoates (PHAs) synthesized by bacteria are the primary source for production of biodegradable bioplastics. PHAs can be produced by microbial cultures fed with renewable feed stocks. PHAs are a great opportunity for the polymer industry. While some bacterial species produce mainly PHB homopolymers consisting only of 3HB monomers, other species can synthesize various PHAs, depending on availability of intermediate precursors. The enzymes synthesizing the polymers, PHA syntheses are encoded by the PhaC genes. Differences in primary and secondary structure determine the capacity to accommodate various hydroxyalkanoate substrates and to form short-chain-length or medium-chain-length PHAs. In this review we discuss the enzyme structure and differences at the base of the characteristic polymers formed. Finally, we review innovative methods applied to biofermentor and on-line fermentation to monitor the production of PHAs, finalized to optimization of fermentation conditions, and the extraction methods available.
Keywords: bacteria, fermentation, feedstock, polyhydroxyalkanoates (PHA), biosensor methods
Polyhydroxyalkanoates (PHAs), synthesized by Gram-negative and Gram-positive bacteria, as well as Archaea and cyanobacteria, are the primary source for production of biodegradable bioplastics. Applications of PHAs are very different and assorted, from coatings, bottles, packaging films, in medical devices and for drug delivery, conductive biopolymers, high-tech electronics, car components, lightweight structural elements in buildings, textile elastomers and fibres, disposable materials, performance additives, transparent films, fertilizer mulches in soil application.1 Poly(3-hydroxybutyrate) (P3HB) is the basic polymer for biodegradable bioplastics, is tough, brittle, fragile, stiff, with low elongation ability, and a break point below 15%. During ageing, PHB tends to re crystallize, and its mechanical properties change with time. PHB is made less stiff through plasticizers addition, and can change mechanical property by adding fibers and compounding materials. Polyhydroxyalkanoate (PHA) polymers contain various 3-hydroxyalkanoate monomers, from 3-hydroxypropionate (3HP), 3-hydroxyvalerate (3HV), 3-hydroxyhexanoate (3HHx), 3-hydroxyheptanoate (3HHp), up to 3-hydroxydodecanoate (3HDD), and in some circumstances also 4-hydroxybutyrate (4HB) and 5-hydroxyvalerate (5HV). PHAs are synthesized in microbial cultures, fed with renewable feed stocks. PHAs are insoluble in water, UV resistant, do not hydrolyze, tend to be degraded in acid and basic solutions, are soluble in chloroform and chlorinated solvents, are biocompatible biologically, being present in human fluids and tissues, are degraded in anaerobiosis, and are not “sticky” during melting, compared to other polymers. Short-chain-length (scl) PHA are formed by 3-hydroxy-C3 to C6 alkane units.2–11 Medium-chain-length (Mcl) PHAs (i.e. composed of 3-hydroxy- C6 to C12 -alkanes) have optimum elongation property, higher break point and lower melting temperature in respect to scl-PHAs.
While some bacterial species produce mainly PHB homopolymers, other species can synthesize PHAs formed by various 3-hydroxyalkanoates, when the required intermediate precursors are provided through engineered metabolic pathways or as feedstock,1 optimized substrate availability (by successive addition of precursors), and use of PHA syntheses with broader substrate requirement, with ability to locate larger hydroxyalkanoate monomers within the substrate binding pocket. The structure and properties of the polymers are largely influenced by the ratio of 3-hydroxyalkanoate monomers in respect to 3-HB. 3-hydroxyvalerate and other co-monomers improve the elasticity of PHAs. The synthesis of copolymers is a frequent strategy to adapt PHAs to the required properties, with improvement of flexibility of bioplastics, that will show lower glass transition temperature (Tg) and melting temperature (Tm).2 PHA homopolymers can have great size, as in high molecular weight (hmw-PHB). Other produced polymers may contain also 4-hydroxybutyrate (P4HB), 3-hydroxy-5-phenylvalerate P(3HPhV), 3-hydroxy-6-phenylhexanoate (3H6PhHs), as in functionalized scl- and mcl- PHAs.
P3HB-4HB polymers are conventional thermoplastic material exploited in packaging. These bioplastics have high tensile strength and high elongation until breakpoint. PHB copolymers containing 3-hydroxyvalerate (P-3HB-co-3HV) have very good mechanical property, being elastic, with good tensile strength, low melting point, and low degree of crystallinity. The thermal, mechanical, and barrier properties of PHBV with different 3HV content, and with various molecular weight, are excellent. The presence of 3HV in the copolymer supports the production process, presents good melting stability at temperatures below 160˚C. P3HB4HB polymers show high tensile strength and higher elongation at break. PHB has been blended with poly(vinyl acetate) (PVAc), two polymers being miscible, achieving good properties and high biodegradability of the products.3 There are several different types of commercially produced PHA polymers, from random copolymers such as poly(3HP-co-4HB), poly(3HB-co-3HP), poly(3HB-co-3HV) (PHBV), poly(3HB-co-4HB) (P3HB4HB), to poly(3HB-co-3HHx) (PHBHHx). Mixed type scl-/mclPHAs, such as P(3HB-co-mcl-3HA), are produced with the NODEX trademark. Production of poly 3-hydroxybutyrate-co-3-hydroxyhexanoate (PHBHHs) has been achieved through the PhaPCJ synthase in Aeromonas hydrophila cultures. Copolymers of P(3HHx-co-3HO-co-3HD-co-3HDD), showing a soft consistence, are also produced. Mixed polyesters containing poly-lactate (pLA), such as poly(LA-co-3HB-co-3HP) are produced industrially.12,13
The PHA syntheses, PhaC, have been classified in four types, depending on the substrate used, and the tendency to produce short chain length (scl-) or medium chain length (mcl-) polymers: type I, type III, and type IV PhaC enzymes synthesize principally scl-PHAs, while type II phaC produce mcl-PHAs.14 PhaC enzymes possess the catalytic triad C-H-D, cysteine, histidine, aspartate, similarly to lipases that prefer serine (S-H-D), and in proteases. The negatively charged aspartate, located at position 447 in PhaCCs from Chromobacterium spp.15 and position 480 in PhaCCn from Cupriavidus necator, enhance the basicity of histidine, through a hydrogen bond: aspartate acts as a general base catalyst, accelerating de protonation of the 3-hydroxyl group in the alkane substrate. The PhaC enzymes differ in the structure of the PhaC, in type I and type II being formed by a monomer that homodimerize, while in type III and IV the catalytic subunit associates to another subunit, forming hetreodimers.
Type I and type II PHA syntheses
Type I and II PHA syntheses are formed by a single protein, about 60 kDa in size. The active PhaC enzyme is a dimer, with catalytic (CAT) domains facing each other, with N-terminal domains making direct contacts, supporting monomer interactions. It was suggested that a partially folded catalytic domain is occupied by a stretch of amino acids (LID) of the CAT domain, able to change conformation and to slide away in presence of activation factors, thus opening the catalytic domain, that become occupied by the 3HB-CoA substrate.14 PhaCCn, a type I enzyme in Cupriavidus necator, is composed of 191 amino acids N-terminal domain, and a C-domain containing the catalytic site, composed of 398 amino acids, for a total of 589 residues (Table 1). In PhaCCn the catalytic triad Cys319, Asp480 and His508, is located between the beta 6 and alpha 3 turn, the beta 10 and alpha 7 turn, and at the end of the beta 11 strand, respectively. Between the Cys319 and the Asp480 there are the Dimerization loop (D-loop) and the helix-turn-helix motif (HTH). PhaCCs from Chromobacterium strains has the property to use either 3HB, 3HV, and 3HH, as substrates, with production of scl-PHAs with mixed composition, and ability to incorporate 3OH-C5 and 3OH-C6 alkanes into the polymer. PhaCCs has a fast polymerization rate, is composed of 560 amino acids (57 kDa in size) but, being shorter amino acid positions change, and compared to PhaCCn. PhaCCs structure has the substrate-binding site hidden by a partially disordered protein domain, the CAP domain, that closes the active site access, with the Cys291 in the catalytic site accessible to substrate when the LID domain slides away.
|
Species |
Type of PhaC |
Catalytic triad: the lipase phaC box serves to form intermediate product Cys-S-H3B |
MW, nr. of amino acids |
Type of polymer |
Mechanism |
|
Cupriavidus necator (Ralstonia eutropha |
Type I (PhaCCn / RePhaC1) |
Cys319, Asp480, His508 |
60 kDa, 589 residues |
Short-chain-length homopolymers (scl-PHA, as PHB) |
helix-loop-helix motif (HTH) and Dimerization loop support dimer formation |
|
Chromobacterium spp., strain USM2 |
Type I (PhaCCs) with mixed substrate range, utilizes 3HB, 3HV and 3HH |
Cys291, Asp447, His477 |
57 kDa, 560 aa, |
scl-PHA polymers with mixed composition |
CAP and LID domain for opening the access and dimerization |
|
Delftia acidovorans (Comamonas acidovorans) |
Type I |
|
61 kDa, 599 aa |
|
|
|
Aeromonas spp. (A. caviae, A. hydrophila, A. punctata, etc.) |
Type I, with mixed substrate range |
|
|
“Nodex”, mixed-type scl-mcl-PHAs |
|
|
Caulobacter crescentus (C. vibrioides) |
Type I, PhaCCc accommodating 3-hydroxyalkanoates with various alkyl side-chain length |
|
57 kDa, 560 aa |
mixed-type scl-PHA |
|
|
Pseudomonas spp. (P. putida, P. mendocina, P. oleovorans, P. campisalis, P. stutzeri) |
Type II, PhaC1, PhaC2 with high affinity for 3HH |
Cys296, Asp452, His453 and His480 (it has been proposed that the first His. His453 is the catalytic histidine) |
|
synthesis of mcl-PHA |
|
|
Halomonas spp., (H. campisalis, Halomonas sp. O-1, H. elongata DSM2581 |
Type II, polymerizing mcl-PHAs |
|
|
mcl-PHAs |
|
|
Chromatium vinosum |
Type III |
|
|
Scl-PHA |
active site cysteine buried in a pocket forming substrate entrance (3OH-alkanoyl-S-CoA), and product exit channel |
|
Thiocapsa pfennigii |
Type III, hybrid Pha synthase |
|
|
Scl-PHA |
easiness of PHB extraction |
|
Haloarcula marismortui |
Type IIIA (archeal) |
|
|
Scl-PHA |
|
|
Bacillus cereus |
Type IV |
C151, Asp306, His335 |
41 kDa |
Scl-PHA |
Alcohol lysis is shared among type IV PHA syntheses: regulation of PHA molec. weight and modification of the PHA carboxyl terminus |
|
B. bataviensis |
Type IV |
|
41 kDa |
Scl-PHA |
|
|
B. megaterium |
Type IV |
C147, Asp302, His331 |
41 kDa |
Scl-PHA |
Table 1 Bacterial species possessing PhaC synthases of different types, and species of PHA produced
PhaCCs due to a change of names with use of Greek letters in the N-terminal secondary structure, there are only six ß-strands (ß8 to ß13) and four a-helices (α4 to α7), whose number is reduced in respect to PhaCCn secondary structure. In PhaCCs the catalytic triad is positioned after the β9 sheet, in respect to the β11 sheet in PhaCCn and Asp447 is located at the ß8-α4 fold, formed by ß8 strand and α4 helix, while in PhaCCn this is the β10-α7 fold. Chuah16 showed by mutation of amino acids that in PhaCCs Ala479 is a critical residue required for substrate specificity, with production of different copolymers, as P(3HB-co-3HHx). In PhaCCn and in other type I enzymes this position corresponds to the conserved residue Ala517. In the study by check, Ala479 is located within α5 helix, with the side chain protruding into a depression of the molecular surface formed by loops β4-α1, β9-α5 and α5-β10 of the core sub domain, and is partially covered by the loop of the LID region: mutation of the A479 resulted in weakening of interactions between the LID region and the core sub domain, and stabilization of the active form of the enzyme, with release of LID region from the catalytic site. Ala479 is supported by polar residues (Ser475 and Arg490), so that mutating Ala479 with Ser or The supports hydrogen-bonding interactions with the polar residues, stabilizing the α5 helix with the His477, part of the catalytic triad. The enzyme PhaCCc from Caulobacter crescentus (C. vibrioides) accommodates 3-hydroxyalkanoates with various alkyl side-chain length. Also in PhaCCc, Ala479 substitution has been reported to increase PhaCCc substrate preference for 3HHx. In Delftia acidovorans the PHA synthase is larger, possessing a 40 amino acid residues stretch that improves the specific activity of the enzyme. Aeromonas species, principally A. caviae, A. hydrophila, and A. punctata, have type I PhaC. These syntheses have mixed substrate specificity, that are exploited in the production of mixed type scl-mcl-PHAs marketedunder the “Nodex” trademark. Type II PHA syntheses are present and distributed in various bacteria: Pseudomonas species contain two phaC genes, of which PhaC1 codify for the active enzyme under physiological conditions. PhaC2 in P. oleovorans has a higher affinity for 3-hydroxyhexanoate (3HH). PhaC1 and PhaC2 in P. stutzeri has been applied to produce mcl-PHAs16–19 in engineered bacteria. PsPhaC2 with four mutated amino acids, E130D, S325T, S477G, and Q481K, was exploiteded to produce mcl-PHAs and block copolymers, being able to host in the active site substrates with various shapes and structures. PhaC syntheses have been described in Halomonas species. Tyr412 in PhaCCs, and Tyr446 in the α6 helix in PhaCCn, are residues conserved in type I, III and IV PHA syntheses, while Phe occupies this position in type II PHA syntheses. In addition, another amino acid showed to have a role in accommodating larger substrates, by means of mutagenesis studies. Tyr438 is conserved in type I, III and IV enzymes, while in type II PhaC this position is occupied by His: the elimination of the bulky side chain of phenol ring can reduce the size of substrate entrance of catalytic site, and change the polar interactions with other amino acids facing the substrate entrance tunnel.
Type III and IV PhaC syntheses
Type III PHA syntheses are composed of two subunits, the catalytic subunit, PhaC (40–53kDa) and a second subunit, PhaE (ranging from 20 to 40kDa), involved in formation of the PhaEC complex, in which PhaE is required for PHA polymerization. Type III PHA syntheses are tetramers, such as in phaEC from Chromatium vinosum. The results of enzyme analysis, referring to PhaCCn and PhaC type III from Chromatium vinosum, describe the presence of the active site in which cysteine is buried in a pocket: Check and coauthors, compared the crystal structure with other enzymes with other enzymes with resolved crystal structure (lipases), and proposed the substrate entrance and product exit channels. An enzyme belonging to type III phaC has been characterized in Thiocapsa pfennigii. There is an Archaeal type, type IIIA PhaC, represented by Haloarcula marismortui. Archea offer promising perspectives of application in bioplastics production, due to the easiness of PHB extraction. Type IV PHA syntheses from Bacillus spp. are composed of the catalytic subunit, PhaC (41.5kDa) and a second subunit, PhaR (22kDa. Type IV PHA syntheses are classified as Bacillus cereus type (IVc), B. megaterium type (IVm), and B. bataviensis type (IVb). In recombinant E. coli expressing PhaRC from B. cereus YB-4, PHA is processed by the synthase alcoholysis and cleavage at the C-terminal. This reaction is exploited to regulate PHA molecular weight and to modify the PHA carboxyl terminal.20
The methods for PHA production in fermentors are industrially important: most production plants make use of patented strains, engineered PHA syntheses, and growth conditions favoring cell high yield and high PHA content/dry cell weight. Recent reviews have described most of the updated processes and applications of bacteria in bioplastics production.21 Bacterial cultures require sterilization; therefore some author proposed the use of halophiles have been proposed to reduce costs of industrial production. Extraction of PHAs from bacteria requires costly processes and methods, that have been described in reviews.1,22,23 Bacteria have been modified to sustain granule formation under high pressure, capable of autolysis in determined conditions.24,25 Researchers proposed the use of Archaea or cyanobacteria that have PHA granules easy to be extracted, to decrease the costs of production.
Biosensors for automatic monitoring of fermentation process
In the monitoring PHA synthesis in biofermentors, various methods have been described, from PHA/lipid staining26 and analysis of fluorescence intensity, to flow cytometry,27 to physicochemical analyses (Raman spectrometry, FTIR, SERS).28 Various detection systems have been applied to quantify the bacteria, with optimum condition for PHA extraction at OD=25-50: the proposed analyses well suited for bacteria concentration evaluation range from spectrophotometric detection, as Fourier Transformation-IR spectrometry (FTIR) and Reflectometric Interference Spectroscopy (RIfS), to Surface Enhanced Raman Spectroscopy (SERS), to electrochemical biosensors, such as Alternate Current (AC) susceptometry. In industrial fermentation for production of PHA, several devices have been proposed to monitor the variables of the process. In particular, the determination of bacteria biomass, the quantification of carbohydrates in the feedstock, and the measure of PHA produced. These control of these parameters can optimize the use of bioreactor for the period of use (reaching the desired bacterial biomass in the shortest time), maximizing the produced PHA at shortest operating times. Various sensors have been applied to biofermentors in PHB production.29 This is a critical point in fermentation, since bacteria that do not synthesise PHAs still consume sugars: often sugars are completely consumed during exponential phase growth so that new feedstock supply is necessary. Traditional PHA screening methods for PHA quantification in whole cells have been based on Nile Blue staining and fluorescence detection.30 Nile Blue dye stains PHB and other neutral lipids, therefore a medium rich in lipids, such as vegetable oils, is easily stained a specifically. New methods have been proposed, such as staining of PHA with lipophilic fluorescent LipidGreen1, and may be run in automatic, providing measures even over-night.26 A quantitative measure of PHA producing bacteria has been based on fluorescence scale reads using a laser scanner.31 (Figure 1) In our hands, the system showed an optimization of feed stock consumption and determination of feed supplementation during exponential growth that occurred contemporary to PHB synthesis, while at the stationary phase determination of maximum PHB presence allowed to stop the biofermentor without further supplementation of feeds. In this way, we obtained an improvement of bacteria growth phase monitoring, in respect to the available systems. Even at low bacterial concentrations, as shown by Optical Density of 2.4 in Figure 1, growth was maximized through evaluation of nutrients requirement, while no nutrient was added when PHB synthesis reached its peak, with containment of materials and costs of biofermentor maintenance. At higher O.D. (i.e. 25 or higher), the system could allow to obtain a high yield of PHB/dry cell content.
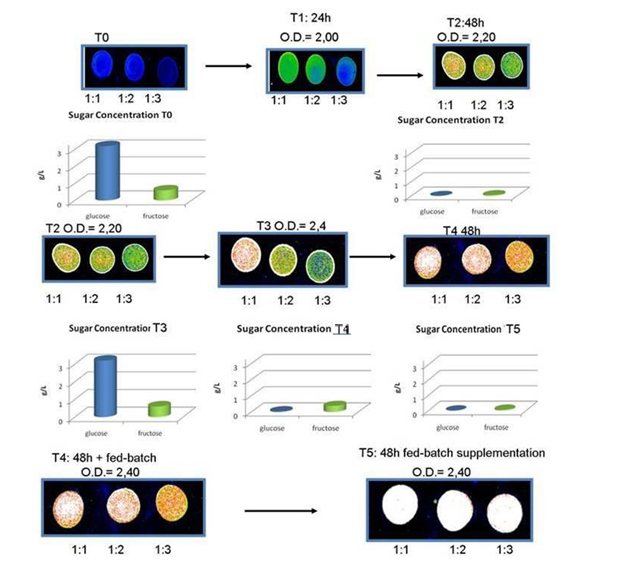
Figure 1 48 hour fermentation in biofermentor ad monitoring of PHB synthesis using Nile Blue staining of bacterial cells and sugars consumption using enzymatic kit. Bacteria were used as total suspensions or diluted 1:2 and 1:3 that were spotted onto glass slides and analyzed on an Affymetrix 428 array Scanner, at excitation/emission 460 nm/550nm. The dilutions showed increase in fluorescence until saturation of signal (white spots). T2 required addition of feed stock, until sugars were totally consumed (T4 and T5).
To the aim of automated detection of PHA in bacteria in fermentors, SERS methods could be effectively applied not only to quantify PHAs, but also to discriminate the type of PHAs produced. This may help to analyze the quality and quantity of PHAs at the same time. The combination of various sensors could make easier the use of bioreactors at industrial scale, optimizing the time of use (bacterial growth) and maximizing the synthesis of PHAs in the shortest time possible. Among the measuring methods or applied to biofermetors, the most favoured are biosensors with few equipment maintenance and less technical involvement. EIS, SPR and other applications require calibration curves, while optical measurements require minimal liquid handling, such as dilution steps and sample replicas. Biofermentors work with high concentrations of sugars in the medium. In the quantification of sugars consumed, and analysis of feedstock, several methods are present based on sticks or test strips, using viscosimetric32 or amperometric detection of glucose oxidase activity. Nano encapsulation of enzymes to read glucose concentration offer also an opportunity for biosensing.33
PHA quantification and extraction
Several approaches have been proposed to make the process economically convenient. For instance, single cell protein method allows fermentation and recovery of granules in a single passage;34–36 there are various lysis methods available,26,37 based on alkaline treatment, before PHA recovery.38 All these protocols are economically convenient and environmentally friendly, not requiring the use of solvents such as chloroform. To measure intracellular polymers, direct cellular digestion in sulphuric acid has been applied. For instance, 50μL of medium are added into 500μL of 96% sulphuric acid in a water-bath at 95-98˚C until PHA is converted to crotonic acid. Spectrophotometric analysis at 235nm is then performed using a quartz cuvette. The PHA concentration is defined as gram of polymer/litre of culture. To extract PHA polymers, cell lysis is obtained by enzymatic digestion. After lysozyme digestion for 1h, a digestion with proteinase K is performed. Then, the digested cell material is transferred in a glass tube. Hot chloroform is added and samples are kept in a water-bath at boiling temperature for 2h (vortexing every 10min). Centrifugation at 3000g for 20minutes allows to remove non-PHB cell material and to recover the lower chloroform phase, containing PHB polymers. Finally, solid PHB/PHA is obtained by non-solvent (methanol and water 7:3vol/vol) precipitation (five volumes for each volume of chloroform) followed by filtration.31
In the near future an increasing interest in industrial production of PHAs with different composition is envisaged. This will attract attention to modified PHA syntheses, engineering biosynthesis pathways, and optimization of fermentation conditions to produce bioplastics at convenient costs.
The activities were performed within the Framework Programme VII of the EU Commission, Collaborative Project targeted to a special Group (such as SMEs) 289603: TRANSBIO: Bio transformation of by-products from fruit and vegetable processing industry into valuable BIO products.
We thank the Prof. Beom Soo Kim and Dr. Prasun Kumar for guest editing a special issue on the journal Polymers that gave us the opportunity to review PhaC syntheses and to collect the material on genes and sequences described in this review.
Authors declare that there is no conflict of interest.

©2018 Poltronieri. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.