International Journal of
eISSN: 2573-2838


Research Article Volume 2 Issue 1
Department of Anatomy, Physiology and Pharmacology, Auburn University, USA
Correspondence: Vitaly Vodyanoy, Department of Anatomy, Physiology and Pharmacology, 207 Greene Hall, Auburn University, Auburn, AL 36849, USA, Tel 334-844-5405
Received: December 04, 2016 | Published: January 23, 2017
Citation: Vodyanoy V. The design of molecular switches for biosensors. Int J Biosen Bioelectron. 2017;2(1):14–18. DOI: 10.15406/ijbsbe.2017.02.00009
A biosensor is a device, which incorporates a biological sensing element near to or integrated with a physical signal transducer. The sensing elements accomplish recognition from the binding, which occurs between bio recognition molecules, and target analytes. Transduction is the physicochemical perturbation caused by this binding that enables recognition of the triggered change by some device. To detect a small number of binding events, a single binding event must be amplified. Here, we design a molecular switch capable of converting a single binding event into the movement of about one million ions per second. In this switch, a single binding, amplified by the release of the stored free electrochemical energy, leads to a dynamic signal that is large compared to the noise in the measuring system. This approach is adapted from biological receptors, which convert chemical signals into currents in ion channels. The molecular switches in this work are artificial ion channels constructed by modular design from molecular pores and gates. The currents through these channels can be registered by conventional methods. The molecular switches can be triggered by various sensing elements such as antibodies, antibody fragments, polypeptides, DNA, RNA, and ion sensitive molecules. The small size and planar architecture of the molecular switches allow them to become components of a microelectronic circuit. The switches can be used for detection of proteins, toxins, viruses, bacteria, and ions.
Keywords: signal transduction, modular design, ion channel, monolayer, receptor, antibody, membrane
In nature, specialized proteins, specific receptors on or in target cells, recognize signaling molecules.1 They specifically bind the signaling molecule and then initiate a response in the target cell. Many receptors use the transduction mechanism utilizing ion channels. These so-called, ion-channel-linked receptors, are also known as transmitter-gated ion channels. The signaling is mediated by binding of a specific molecule, which that opens or closes the gate of the ion channel, thereby changing the ion permeability and electrical conductance of the channel.
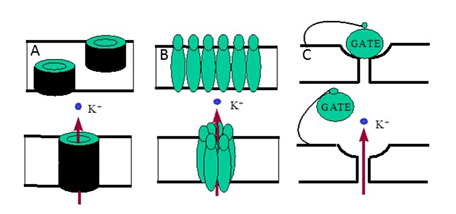
Thus, these transmitter-gated ion channels work as chemo electric transducers. There are several molecular mechanisms of opening and closing of natural and artificial ion channels.2 Channels can be opened or closed as a result of aggregation – disaggregation of molecular subunits constituting a channel (Figures 1A & 1B) or, a molecular gate can control the closing and opening of the channel (Figure 1C). A model system with an antibiotic, gramicidin, where two half-channels connect up, was used to construct a biosensor.3 The “off” state of the channel was realized by the displacement of one-half of the channel due to binding of the analyte.
The chief drawbacks of the gramicidin switch were the need for the displacement of the relatively large gramicidin molecule for switching and the absence of independent gate. Genetically engineered protein pores4 provide another approach to making molecular switches for biosensors. Protein pores could provide speed and sensitivity as transducers. However, these relatively large protein molecules are difficult to maintain outside the living cell.
Modular design
The task of this article is the design of a molecular switch capable of amplification and transduction of a chemical signal into an electrical current to be registered by a conventional electronic instrument. Pore-forming polyene antibiotics and synthetic channel gates immobilized in the artificial supported planar membrane are to be used to compose a modular molecular system capable of switching ion channels mediated by specific binding. Two molecular models are considered for practical applications in biosensors. One molecular switch is intended to be in the NORMALLY “ON” state, which can go to the “OFF” state when a specific molecule is bound to a trigger site. Another switch has a NORMALLY “OFF” state that goes “ON” after a specific binding. In the “ON” state the channel is open and ions can go through, while in the “OFF” state the channel is shut off and ions are stopped.
Requirements
Design an artificial molecular switch capable of performing amplified signal transduction between a variety of chemical sensing elements and an electronic device. The switch needs to be sensitive, fast, and suitable for practical biosensors.
Approach
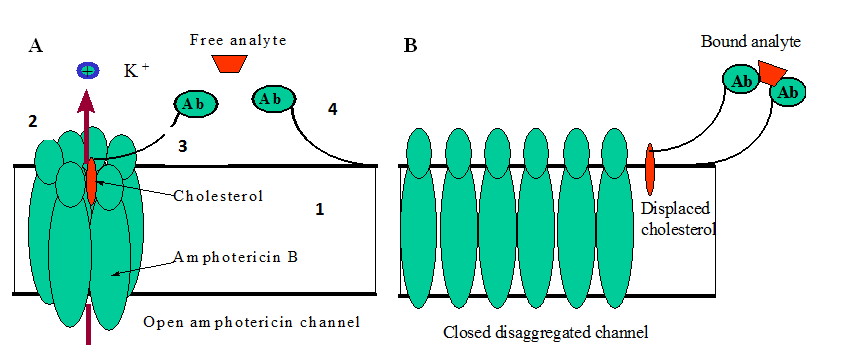
For the NORMALLY “ON” switch we use molecules of the polyene antibiotic amphotericin B (AmB) known to form ion channels in artificial lipid membranes.5-9 The presence of the cholesterol molecules in the AmB channel is critical for the channel to be open.10 The removal of the cholesterol from the channel shuts the channel off. Thus, the switch, in this case, is the modular assembly of the amphotericin pore and the cholesterol gate tethered with the sensing element (antibody, polypeptide, etc.). The channel is switched when the target is recognized, bound to the sensing element, and the displacement of the cholesterol gate shuts the channel off.
The major modular components of the NORMALLY “ON “molecular switch (Figure 3) are phospholipids for the supporting membrane (Figure 1 & 3A), amphotericin B for construction of the ion channel (Figure 2 & 3A), the cholesterol gate (Figures 3A, 3 & 4), and the molecular anchor (Figure 3A) ( Figure 4). Phospholipids and amphotericin B are commercially available from Avanty Polar Lipids, Inc. and Sigma Chemical Co. The displaceable cholesterol gate is composed of the cholesterol molecule linked to an antibody (SynPep CA). The molecular anchor is the same antibody linked to the supporting membrane through stearic acid (SynPep, CA) or biotinylated lipid and streptavidin (Molecular Probs Inc, OR). For the proof of concept, the 7-base peptide specific to skeletal muscle protein as an antibody can be used.11,12
The cholesterol molecule holds the aggregated channel open by zipping the amphotericin molecules together (Figure 3A). When the channel is open, ions can pass through. When the analyte binds the antibody, it displaces the cholesterol and unzips the channel. This results in disaggregation and closing of the channel (Figure 3B). The process is reversible. When an antibody is unbound, the liberated cholesterol can again zip the channel open. The NORMALLY “OFF” switch is constructed as a modular assembly of a small artificial channel and a synthetic molecular gate tethered to the base of the supporting membrane. The channel opens when the target binds the sensing molecules and pulls the plug out of the channel.
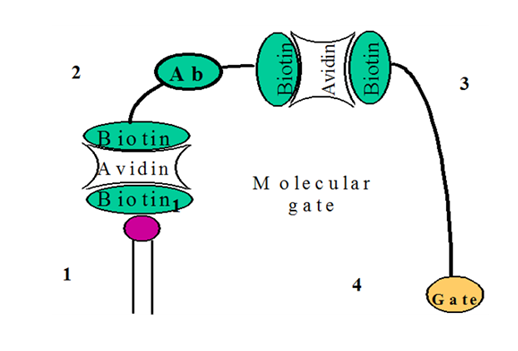
The major modular components of the NORMALLY “OFF” molecular switch are phospholipids for supporting membrane (Figure 1) (Figure 5A), the amphotericin B and cholesterol for construction of the ion channel (Figure 2) (Figure 5A), the synthetic molecular gate (Figure 3) (Figure 5A), and molecular anchor (Figure 4) (Figure 5A). The supporting membrane, the open amphotericin B-cholesterol channel, and the molecular anchor are the same as in the NORMALLY “ON” switch. The displaceable cholesterol gate is unused for this switch. Instead, we use a synthetic molecular gate. We use a tethered blocker of K+ -ions (ion plug), attached to a sensing element and membrane with a flexible molecular linker. Components of the molecular gate are shown in Figure 7. The biotinylated lipid (Figure 1) (Figure 6) and the sensing peptide (Figure 2) (Figure 6) are the same as in the NORMALLY “OFF” switch. The biotinylated channel blocker is constructed by using an extender arm maid of ethylene glycol linkages (Figure 3) (Figure 6) and anion, gate (Figure 4) (Figure 6). Extender arms are to be constructed from the spacer-linked biotins available commercially. The gates are to be constructed from known potassium ion blockers 2 and spacer-link biotinylated compounds (Table 1) (Table 2).
Compound |
Length of Linker |
Source |
aSulfo-NHS-LC-Biotin |
12 atoms |
Pierce |
bBiotin-XX-NHS |
13 atoms |
Calbiochem |
cBiotin-TEG |
15 atom3 |
Pierce |
dSulfo-NHS-LC-LC-Biotin |
19 atoms |
Pierce |
Table 1 Examples of spacer-link biotinylated compounds
aAmine-reactive biotinylation reagent
bN-Hydroxysuccinimide (NHS)
cBiotin TEG: 15 atom triethylene glycol spacer
dAmine-reactive biotinylation reagent, double long chain
Tetraethylammonium |
4-aminopyridine |
Apamin |
|
|
|
Table 2 Selected K+ ion channel blockers
For example, 4-aminopiridine and apamin gates are to be constructed using the reaction of the Sulfo-NHS-LC-LC-Biotin (water soluble) with 4-aminopiridine and apamin, respectively, making the amide bond with the linker

Immobilization of modular components of the molecular switch
A critical step in the development of the molecular switch is the immobilization of the modular components, to the platform area on the chip where the target molecules will bind in a positive response. Three methods are used to immobilize molecules on the sensor surfaces1 Langmuir-Blodgett (LB) deposition of a composite lipid monolayer onto a sensor surface2,6,13 antibody-liposome conjugation14 and3 molecular self-assembling using biotin/streptavidin coupling.15 The precise thickness of LB films, coupled with the degree of control over the molecular architecture, has now firmly established a role for these organic multi layers in thin film technology. Also, LB and molecular assembling technologies can provide a way to produce biocompatible hydrophilic (or hydrophobic) surfaces useful for adsorption of various capture ligands. The LB method uses monolayers containing ligands with high specificity and low non-specific binding. As result, a dynamic equilibrium (association-dissociation) of antibody-antigen interaction can be rapidly achieved and measured. The monolayer in this method is formed on the air-liquid interface by allowing the spreading solution to run down a inclined wettable planar surface that is partially submersed into the sub phase. Membrane vesicles are positioned on the wet slide at the edge of positive meniscus of liquid, at the liquid-air interface. The modular components of the molecular switch are bound to the vesicular membrane. When surface forces rupture the vesicle and it splits into a monolayer the unique process of purification occurs: membrane-bound molecules are left bound to the newly created monolayer, but soluble impurities go into the sub phase beneath the monolayer. When the monolayer is compressed and transferred onto sensor surface there are only membrane-bound molecular components surrounded by compatible lipids. The spreading material is purified in process of spreading, it needs no special preliminary purification. The method is simple and cost-effective.
Monolayer formation and deposition for molecular devices
Phospholipid monolayers: Phospholipid solutions are spread on the surface balance as hexane solutions (1 mg/ml) containing 2% ethanol. The sub phase used in the experiments is a solution containing 55 mM KCl, 4 mM NaCl, 0.1 mM CaCl2, 1 mM MgCl2 and 2 mM 3-(N-morpholino)-propanesulfonic acid (MOPS) made with de ionized doubly distilled water (pH adjusted to 7.4 with KOH).
Monolayers made of antibody preparations: The quartz crystals covered with gold are cleaned by treatment with 50 % (v/v) HNO3 and are rinsed in running distilled water until the acid is completely removed. The crystals are then dried and stored until further use. The monolayer is formed on the air-liquid interface of Lngmuir-Blodget system by allowing the spreading solution to run down a inclined wettable planar surface that is partially submersed into the sub phase. 150 ml of the antiserum is spread on the sub phase surface by allowing it to flow down a wet glass plate that crossed the interface. The flow rate down the plate is maintained about 0.1 ml/min. After spreading the glass plate is removed, and the monolayer is allowed to equilibrate and stabilize for 10 minutes at 19 ± 0.1 0C. The monolayer is then compressed at a rate of 30 mm/min and the vertical film deposition is carried out with a vertical rate of 4.5 mm/min and at a constant surface pressure of 23 mN/m. Seven monolayers of the antibody (phage, peptide, virus) film are transferred to the gold surface of the quartz crystals in this manner. Monolayers containing antibodies are transferred at a constant surface pressure onto a silicon/silicon dioxide plate, or onto standard microscope slides for visual observations and cell counting. Silicon/silicon dioxide plates (10´10 mm2) with etched silicon windows of 2´2 mm2 in the center of the plate are used as sensor surfaces(Table 3).
Features of the Molecular Switch |
High Sensitivity from 1 To 100 Binding Events |
Gain 1´106 |
Real Time Operation |
Modular Molecular Design |
Can be Triggered by a Large Variety of Sensing Elements |
Reversible |
Electrical Output Signal |
Small Size and Planar Architecture |
Table 3 Features of the Molecular Switch
Advantages of the molecular switch
Estimate of amplification of the molecular switch
The binding of a single analyte molecule can open a molecular switch and facilitate a flux of one million ions per second. Let us consider the momentary binding of specific target molecules to antibodies on the surface of a membrane. If for example, the binding molecules are amino acids, then the time constant of the adsorption is 10-4-10 sec. The highest electrical charge each of them can bring to the interface is 1.6´10-19C. The average charging current is I=dQ/dt=1.6´10-15-1.6´10-20A. If the binding of the target molecule causes the molecular switch to open an ion channel of 10-100 pS at 100mV, this will result in a channel current Ic=10-12-10-11A. In that case, the average amplification coefficient, Ka=Ic/I, can be estimated as 106. The measurement of single ion channel currents is the known technology.2
Application of molecular switches
The single-molecule selectivity and specificity of the molecular recognition together with the high amplification have a promise to be utilized in many applications.16 Sensitive and molecular switches can be used for high-throughput screening of, drugs, environmental monitoring, food safety, and biowarefare control.17 Molecular switches can be used for detection of multiple pathogens.18 The study of molecular switches is expected to lead to the development of new sensors for a wide range of molecules.
We have designed a molecular switch capable of converting a single binding event into the movement of about one million ions per second. In this switch, a single binding, amplified by the release of the stored free electrochemical energy, leads to a dynamic signal that is large compared to the noise in the measuring system. The molecular switches in this work are artificial ion channels constructed by modular design from molecular pores and gates. The molecular switches can be activated by antibodies, nucleic acids, peptides, and ion sensitive molecules.
Supported by grants from DARPA, MDA972-00-1-0011 and NIST, 70 NANB14H324
The author declares no conflict of interest.

©2017 Vodyanoy. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.