eISSN: 2471-0016


Case Report Volume 3 Issue 3
1Department of Pathology, St Antonius Hospital, Netherland
1Department of Pathology, St Antonius Hospital, Netherland
2Department of Pathology, University Medical Center, Netherland
2Department of Pathology, University Medical Center, Netherland
3Department of Pulmonary Diseases, Rijnstate Hospital, Netherland
3Department of Pulmonary Diseases, Rijnstate Hospital, Netherland
4Department of Pulmonary Diseases, St Antonius Hospital, Netherland
4Department of Pulmonary Diseases, St Antonius Hospital, Netherland
Correspondence: J Alain Kummer , St. Antonius Hospital, Department of Pathology, PO Box 2500, 3430 EM, Nieuwegein, the Netherlands
Received: January 01, 1971 | Published: December 28, 2016
Citation: Smits AJJ, Roepman P, Claessens NJM, et al. Lung adenocarcinomas: a novel KRAS/EGFR exon 21 double mutation with limited response to TKI treatment and two rare EGFR exon 19 deletions with variable response. Int Clin Pathol J. 2016;3(3):225–228. DOI: 10.15406/icpjl.2016.03.00078
EGFR, epidermal growth factor receptor; TKIs, tyrosine kinase inhibitors; PFS, progression-free survival
Lung adenocarcinoma patients whose tumors harbor activating mutations of the Epidermal Growth Factor Receptor (EGFR) gene can benefit from treatment with small-molecule EGFR tyrosine kinase inhibitors (TKIs) like gefitinib and erlotinib. In-frame deletions in exon 19 (usually 15 or 18bp) and the exon21 point mutation Leu858Arg account for approximately 90% of EGFR mutations.1 These specific types of mutations have been shown to confer increased sensitivity to TKI treatment in multiple studies, leading to higher response rates and longer progression-free survival (PFS) compared to standard chemotherapy.2–4 The clinical implications of EGFR TKI treatment for other very rare EGFR mutations are less clear. Since the frequency of these mutations is so low the sensitivity to TKI treatment cannot be tested in controlled trials and therefore there was a recent encouragement in the Journal of Thoracic Oncology to submit data concerning clinical response to TKI treatment on individual case basis.5 Here, we present a case of a novel EGFR exon21 point mutation combined with a common activating KRAS mutation, and two cases of rare EGFR exon19 deletions.
Case 1
A 57-year-old white male smoker (number of pack years unclear) with a history of peripheral vascular disease and alcohol abuse presented in September 2013 with stage IV (cT2aNoM1b) lung adenocarcinoma. There were pleural, osseous, and cerebellar metastases. EGFR/KRAS mutation analysis was performed by Sanger sequencing on a biopsy of the iliac bone. This revealed a novel EGFR exon 21 missense mutation c.2621G>A (p.Gly874Asp), combined with a KRAS exon 2 missense mutation c. 34G>T (p.Gly12Cys).
First-line chemotherapy with cisplatin/pemetrexed was started. There was disease progression after two rounds of chemotherapy and second-line treatment with gefitinib 250 mg/day was started. After two months there was progression of the brain metastases, for which whole brain radiotherapy was given, combined with palliative radiotherapy on the hip. TKI treatment was interrupted for one week and continued after the radiotherapy. Chest X-rays showed stable disease of the primary tumor (maximal response 10% tumor shrinkage). After 5 months there was extensive disease progression with an increase in size of the primary tumor and multiple liver metastases. The therapy was changed to best supportive care and the patient died a few days later.
Case 2
A 66-year-old white female without relevant medical history and without a history of smoking presented in June 2014 with a tumor in the left upper lobe. Clinical workup showed cT1bN2M1b cancer (with one liver metastasis and multiple bone metastases). A diagnosis of metastatic pulmonary adenocarcinoma was confirmed in a liver biopsy. The tumor cells were positive for CK7 and TTF-1. EGFR/KRAS mutation analysis was performed by high resolution melting analysis (HRM) followed by Sanger sequencing. In EGFR exon 19 a deletion of nucleotides 2236 through 2251 and insertion of one nucleotide (T) was detected (c.2236_2251delinsT). This gives rise to replacement of amino acids 746 through 751 (Glu-Leu-Arg-Glu-Ala-Thr) by a Serine residue (p.Glu746_Thr751delinsSer). KRAS codons 12, 13, and 61 were wild type.
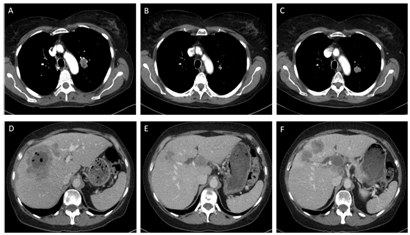
A treatment with erlotinib 150 mg/day was started in July 2014 and imaging in September 2014 showed partial response (Figure 1). In October 2014 a new metastasis in the liver was detected, which was treated with stereotactic radiotherapy in November 2014. Since the other lesions were stable, treatment with erlotinib was continued until there was progression of the primary tumor and liver metastases in February 2015 (Figure 1).
Case 3
A 64-year-old white male with a history of heavy smoking until 2009 (90 pack years) and without other relevant medical history presented in May 2014 with a tumor in the right lower lobe. Clinical workup showed cT2aN2M1a metastatic cancer and the diagnosis of pulmonary adenocarcinoma was confirmed on a lung biopsy. The tumor cells were positive for CK7 and TTF-1. EGFR/KRAS mutation analysis was performed by HRM followed by Sanger sequencing. In EGFR exon 19 a deletion of nucleotides 2236 through 2253 and insertion of three nucleotides (ATT) was detected (c.2236_2253delinsATT). This gives rise to replacement of amino acids 746 through 751 (Glu-Leu-Arg-Glu-Ala-Thr) by an Isoleucine residue (p.Glu746_Thr751delinsIle). KRAS codons 12, 13, and 61 were wild type.
Treatment with gefitinib 250 mg/day was started in May 2014 together with talc pleurodesis and follow up imaging showed a partial response, still ongoing in November 2014. The most recent follow up in March 2015 showed stable disease and gefitinib treatment was continued.
EGFR mutation positive lung adenocarcinoma patients can benefit from TKI treatment, which not only leads to longer PFS compared to standard chemotherapy,2-4 but also to improved quality of life.6 The relationship with these treatment outcomes, however, has only been firmly established for the most frequent types of EGFR mutations, while there is limited or no information concerning many rare mutations.
Here we describe two patients with lung adenocarcinomas harboring rare EGFR exon 19 deletion/insertion mutations, one of whom showed tumor progression after 3 months of erlotinib treatment, while the other patient has stable disease after 9 months of gefitinib treatment. The c.2236_2251delinsT mutation in patient #2 has been described once before, but this reported case was not treated with TKIs.7 The c.2236_2253delinsATT mutation in patient #3 has never been described at the DNA level, but six cases with the same mutation at the protein level (p.Glu746_Thr751delinsIle) have been described.8-13 In three of these studies patients have been treated with TKIs. There is one case of a c.2235_2252delinsAAT mutation with partial response to TKI treatment (gefitinib or erlotinib not specified)13 and one case (not specified at the DNA level) with complete response to treatment with erlotinib.12 In the third reported case (c.2236_2252delinsAT) it is unclear whether this patient was or was not treated with gefitinib, because the treatment is not specified per individual patient and not all patients in the study were treated with TKIs.10
EGFR and KRAS mutations are generally considered to be mutually exclusive,14,15 although some reports of combined EGFR and KRAS mutations exist.16-18 The association between activating mutations in codons 12 and 13 of KRAS and resistance to TKI treatment has been demonstrated in 2008.19 While combined EGFR and KRAS mutations are very rare, reports concerning TKI treatment of these patients are even rarer and the described response to treatment is variable, but at least in cases with an EGFR mutation that by itself is known to be responsive to TKIs, response has been described in patients with combined mutations.16,18
In the case of patient #1, the EGFR mutation was novel and thus responsiveness to TKIs had not been reported. In this patient the primary tumor showed a short-lasting response (for a period of 5 months), but the brain metastases showed rapid progression (after 2 months already). This could be due to limited permeability of the blood brain barrier to TKIs20 or to a difference in EGFR mutation status between tumor locations.21,22 According to a publication by Jackman et al,23 patients who experience progression of central nervous system lesions only should not be considered as having systemic acquired resistance to TKIs,23 but the revised Response Evaluation Criteria in Solid Tumors (RECIST 1.1) do not make an exception for these cases when it comes to the definition of progressive disease. 24
The three patients presented here show that the response to TKI treatment of tumors carrying rare EGFR mutations can be variable, as has also been reported by others .25 Due to the very low frequency of rare EGFR mutations, it is not realistic that the response to TKI for these specific cases will be investigated using a randomized controlled trial. Instead we need to rely on reported case studies in the literature and entries in databases, such as the Catalogues of Somatic Mutations in Cancer (COSMIC) database.26 For unknown or unreported mutational cases we can try to predict the response in silico by emerging bioinformatic approaches including functional predictive models such as Poly Phen27 and SIFT,28 but possibly better by models that predict specific 3D structures of EGFR mutant molecules and the effect on binding of gefitinib or erlotinib.29,30
We retrospectively investigated the usefulness of this information for the three cases reported here (Table 1). For comparison, we also added the information regarding the well-known responsive Leu858Arg and resistant Thr790Met mutations. Unfortunately, for the novel EGFR mutation of patient #1 at codon 874 no data were yet available in the EGFR structural database.29 For patient #2 the precise mutation was also not present in the database, but information regarding erlotinib binding efficiency was provided for a mutation that was closely related (p.Glu746_Thr751delinsSer). Based on the lower binding energy it was predicted to indicate a responsive mutation, similar to the Leu858Arg mutation. However, the patient only showed a limited partial response with a new liver metastasis within 2 months after starting with gefitinib treatment. As such, we believe that the binding energy data were not representative for the actual p.Glu747_Thr751delinsSer mutation in which the exon 19 deletion comprised one less amino acid. For patient #3 the observed mutation (p.Glu746_Thr751delinsIle) was present in the database with a relatively low binding energy (-42.2) indicating a mutant EGFR that is likely to respond to TKI . Indeed this patient showed stable disease for almost a year until the last follow-up in March 2015.
Patient |
KRAS Status |
EGFR Status |
Reported in COSMIC* |
Predicted Effect# |
Observed TKI Benefit |
#1 |
c.34G>T (p.Gly12Cys) |
c.2621G>A |
No |
Not available |
Disease progression |
#2 |
wildtype |
c.2236_2251delinsT |
1 |
-44.3 |
Partial response |
#3 |
wildtype |
c.2236_2253delinsATT |
no (6 similar on protein level) |
-42.2 |
Stable disease |
|
|
Leu858Arg |
|
-46.0 |
Responsive |
|
|
Thr790Met |
|
-36.4 |
Resistant |
Table 1 Summary of reported EGFR/KRAS mutational cases.
*Search data COSMIC database: 09-18-2015.
#Prediction based on data provided in the EGFR mutant structural database.
(http://bcc.ee.cityu.edu.hk/data/EGFR.html). A lower binding energy indicates a better binding of the EGFR mutant molecule and the TKI, suggesting a better inhibition by the drug [29].
For the cases reported here it remained difficult to predict the response to TKI using in silico models and databases. However with the quickly developing field of bioinformatics and the great increase in available data of lung adenocarcinoma mutations this will likely improve in the future. For now we mostly need to rely on information in reported case studies and, importantly, on discussions between molecular biologists, pathologists, and clinicians to determine the best treatment strategy, including an active feedback from the clinic to the diagnostic department whether the patient responded to the selected treatment.
None.
The author declares no conflict of interest.

©2016 Smits, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.