eISSN: 2576-4497


Research Article Volume 1 Issue 2
1Oncology department, Alexandria University, Egypt
2Oncology department, Mansoura University, Egypt
3Department of molecular biology, Alexandria University, Egypt
4Professor of Developmental Genetics, Alexandria University, Egypt
5Department of molecular biology, Alexandria University, Egypt
6Department of biology, National research center Cairo, Egypt
Correspondence: Waleed Arafat, Assistant professor of oncology, Alexandria university, oncology department, Fellow of MD, Anderson Texas university, Fellow of UAB, Birmingham, Alabama, USA, Tel (+2)01223399328
Received: May 21, 2017 | Published: June 14, 2017
Citation: Arafat W, Ashry MSE, Alrazek MAA, et al. The relation between survivin gene expression and urinary bladder cancer disease. Hos Pal Med Int Jnl. 2017;1(2):26–33. DOI: 10.15406/hpmij.2017.01.00007
Purpose: The aim of this study is to investigate the expression of survivin gene in patients suffering from urinary bladder cancer, thus contributing to further understanding of the molecular etiology of the disease by assessment of Survivin gene expression on both RNA and protein levels in urinary bladder tissue samples and Correlate between its expression and pathological prognostic parameters of patients with urinary bladder cancer risk.
Methods: In the present study 26 cases of urinary bladder cancer and 26 cases of normal tissues far from tumor sites in the urinary bladder of the same patients (as control samples) were collected, also 10 paraffin-embedded tissue samples from consecutive of archival bladder specimens patients and 10 tissue samples that of healthy tissue from the same individual as control group were collected for immunohistochemical detection of survivin protein and real time PCR for survivin mRNA detection.
Results: In normal tissues of control samples no expression for survivin gene has been noticed. However, in patients affected with bladder cancer the survivin expression was highly increased by increasing degree of tumor malignancy as indicated by real time PCR for survivin mRNA and immunohistochemistry for survivin protein.
Conclusion: From this study, it is concluded that survivin protein was not expressed in normal bladder urothelium but was present in a high percentage in tissue samples of patients suffering from bladder cancer and its expression is associated with disease recurrence, tumor grade, progression and mortality.
Bladder cancer is the 9th most common cancer diagnosis worldwide, with more than 330,000 new cases each year and more than 130,000 deaths per year, with an estimated male:female ratio of 3.8:1.0 In industrialized Western countries, transitional cell tumors comprise 90%-95% of bladder tumors; 3%-7% are squamous cell, and 1%-2% are adenocarcinomas.1–3 In developing countries in certain locations, up to 75% of cases are squamous cell carcinomas associated with Schistosoma haematobium infestation.2 In Egypt, bladder cancer has been the most common cancer during the past 50 years. Previous research has reported a significant decrease in SCC in Egypt, although the overall bladder cancer incidence in Egypt has remained steady due to an increase in TCC over the past 30 years.4–6 Over the last few decades, there have been advances in urinary bladder cancer management leading to earlier detection of the disease and the development of more effective treatments, improving the outcomes for people living with the disease and resulting in declines in urinary bladder cancer deaths.7–9 However, recent studies in molecular biology and genetics have identified survivin as a candidate gene responsible for cancer progression.10
Survivin, a unique and the smallest member of the Inhibitor of Apoptosis (IAP) family, also called baculoviral inhibitor of apoptosis repeat-containing 5 or BIRC5, is a protein that, in humans, is encoded by the BIRC5 gene. The survivin gene on chromosome 17q25 is now believed to consist of 4 dominant exons (1,2,3 AND 4) and 3 additional cryptic exons located either in intron 2(2alpha_, 2B) or intron 3(3B) and is required for cell division.11–12 It was demonstrated that the functions of survivin in cell division fall into three main categories: participation in chromosomal passenger protein biology, participation in the spindle checkpoint, and regulation of microtubule dynamics.13,14 The ability of transformed cells to evade apoptosis and favor cell survival is a hallmark of tumor progression and resistance to therapy.14 Understanding the multiple functional roles of survivin in cancer, including: resistance to cancer therapy, promoting angiogenesis and preventing cells from apoptosis makes this a relevant target protein for cancer biologists to comprehend.15
Survivin is also involved in the resistance of tumor cells to both chemotherapy and ionising irradiation, that’s why better understanding of survivin and its role in bladder cancer will help better understanding of the molecular biology of the disease and improvement of treatment.116–18 In this study we aim to investigate Survivin gene expression on both RNA and protein levels in urinary bladder tissue samples and Correlate between survivin expression and pathological prognostic parameters of patients with urinary bladder cancer risk.
Patients
All subjects were recruited from the Urology and Nephrology Center Mansoura University and Alexandria university hospital - Egypt.
The present study was conducted on a total of 26 Egyptian individuals:
Immunohistochemical staining for survivin was done using the procedure (According to: Buchwalow and Bocher, 2010) and Immunohistochemical staining protocol (Genemed, USA):
The list of reagents used for Immunohistochemical staining and mRNA extraction process could be found in (Table 1).
Reagents for Immunohistochemical Staining for Survivin Protein |
Reagents for RNA Extraction and cDNA Synthesis Used in Real Time PCR Detection of Survivin MRna |
For immunohistochemistry staining:
|
For real time PCR Formulation of sequence-specific primers (according to Asanuma et al. [19])
|
Table 1 The list of reagents used in this study.
The procedure included
The Immunohistochemical staining protocol (Genemed, USA) included:
Detection of survivin mRNA expression by real-time PCR
Procedure For RNA extraction from 26 tumor and 26 control tissues was done according to Chomczynski and Sacchi, 1987. This step was followed by cDNA synthesis and the reverse transcription reaction product can be directly used in PCR applications or stored at -20°C for less than one week. For longer storage, -70°C is recommended.
For real time PCR using TaqMan probes (Figure 1): According to Asanuma et al.19 quantitative RT-PCR was performed using a TaqMan Universal PCR Master Mix (Applied Biosystems, USA). The reaction mixture (50µl final volumes) contained 5µl of cDNA solution, 25µl of 2X Master Mix, 200nM forward and reverse primers, and 100 nM probe. The amplification cycles were 95°C for 15 seconds and 60°C for 1 min, and repeated for 40 cycles. The quantity of specific mRNA was normalized as a ratio to the amount of GAPDH, which was determined using TaqMan® GAPDH Control Reagents kit (Applied Biosystems, USA), according to manufacturers protocol.
Statistical methods
The spss ANOVA was used to test the difference between normalized survivin values and disease-developing categories.
Different clinicopathological features among studied population
In the studied group we assessed male to-female ratio, tumor grade, Pathologic Types, lymph node metastasis and tumor size according and stage of disease according to the TNM 2009 classification (Table 2). However, the follow up of these patients to detect the survival and treatment outcome will be assessed in further future studies.
Clinicopathological Features |
Total (n=26) |
|
No. |
% |
|
Age |
||
31-50 |
4 |
15.38% |
51-70 |
19 |
73.07% |
71-90 |
3 |
11.53% |
Range |
36-79 |
|
Sex |
||
Male |
22 |
84.61% |
Female |
4 |
15.38% |
Mean |
58.96 |
|
Median |
59 |
|
S.D |
9.45 |
|
Histological Types |
||
Adenocarcinoma |
2 |
7.69 |
Transitional cell carcinoma |
22 |
84.61 |
Small cell carcinoma |
1 |
3.85 |
Squamous cell carcinoma |
1 |
3.85 |
Grade |
||
Grade I |
5 |
19.23 |
Grade II |
5 |
19.23 |
Grade III |
16 |
61.54 |
Stage |
||
Stage I |
2 |
7.96 |
Stage II |
8 |
30.77 |
Stage III |
9 |
34.61 |
Stage IV |
7 |
26.9 |
Size |
||
< 5 |
10 |
38.5 |
5 - 10 |
13 |
50 |
> 10 |
3 |
11.5 |
Range |
1.5 - 12 |
|
Lymph Node Status |
||
0 |
19 |
73.08 |
1 |
2 |
7.69 |
2 |
5 |
19.23 |
Table 2 The distribution of different Clinicopathological features among studied population.
Molecular results
Tissue expression of survivin mRNA quantitative survivin mRNA expression was measured in specimens excised from bladder cancer patients with varying stage and grade categories. In addition healthy tissue specimens from the same study individuals as a negative control group. RT-PCR analysis revealed sufficient expression in all cancer investigated samples. Representative data obtained in cancer specimens are shown in Figure 2.
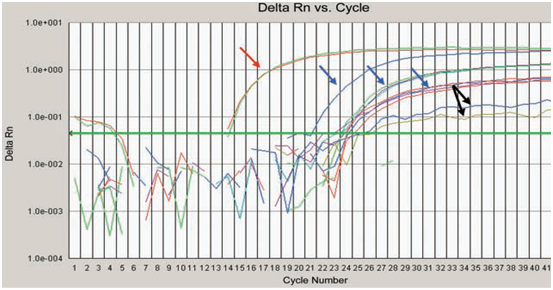
Figure 2 Amplification curves of surviving mRNA extracted from bladder cancer patients. The last two curves indicated by black arrows are samples represent low expression of survivin gene.
Characteristics |
No. |
% |
Relative Expression of Surviving mRNA in Bladder Cancer Patients |
||||
Mean |
Std. Deviation |
Minimum |
Maximum |
||||
Grade |
|||||||
Grade1 |
5 |
19.23 |
64.492 |
54.93405 |
4.28 |
128 |
|
Grade2 |
5 |
19.23 |
641.648 |
975.6593 |
32 |
2352.53 |
|
Grade3 |
16 |
61.54 |
1684.398 |
1307.671 |
147.03 |
4096 |
|
F |
4.581 |
||||||
Stage |
|||||||
Early stage |
10 |
38.46 |
802.08 |
652.5 |
32 |
4096 |
|
Advanced stage |
16 |
61.54 |
1403.78 |
865 |
4.28 |
4096 |
|
p |
0.04* |
||||||
Lymph Node Status |
|||||||
N0 |
19 |
73.08 |
894.8 |
1129.7 |
4.3 |
4096 |
|
N1 |
2 |
7.69 |
1806.6 |
772 |
1260.7 |
2352.5 |
|
N2 |
5 |
19.23 |
2141 |
1642.3 |
147 |
4096 |
|
F |
1.802 |
||||||
Histological Tumor Types |
|||||||
Adenocarcinoma |
2 |
7.69 |
384 |
181.0193 |
256 |
512 |
|
SCC |
1 |
3.85 |
9.18 |
. |
9.18 |
9.18 |
|
Small cell carcenoma |
1 |
3.85 |
90.5 |
. |
90.5 |
90.5 |
|
TCC |
22 |
84.16 |
1346.063 |
1322.818 |
4.28 |
4096 |
|
Total |
26 |
100 |
1172.349 |
1283.897 |
4.28 |
4096 |
|
F |
0.883 |
||||||
Table 3 The correlation between survivin mRNA expression and different bladder cancer pathological characteristics.
Immunohistochemical results (Table 4)
(From 10 paraffin-embedded tissue samples)
Survivin protein was overexpressed in urinary bladder cancer tissues. Positive or negative survivin overexpressed was determined by IHC staining. Cells that had positive survivin overexpression, revealed with brown nuclei, where cells that had negative survivin overexpression, revealed with blue nuclei. The intensity of the staining was designated as
Pathological Features |
Gene Expression |
Total |
|||||||
+ |
++ |
+++ |
|||||||
Grade |
|||||||||
Grade |
I |
No |
4 |
0 |
0 |
4 |
|||
% |
100.00% |
0.00% |
0.00% |
40.00% |
|||||
II |
No |
0 |
2 |
0 |
2 |
||||
% |
0.00% |
100.00% |
0.00% |
20.00% |
|||||
III |
No |
0 |
0 |
4 |
4 |
||||
% |
0.00% |
0.00% |
100.00% |
40.00% |
|||||
Total |
No |
4 |
2 |
4 |
10 |
||||
% |
100.00% |
100.00% |
100.00% |
100.00% |
|||||
P = 0.0001* |
|||||||||
Stage |
|||||||||
Stage |
I |
No |
3 |
0 |
0 |
3 |
|||
% |
75.00% |
0.00% |
0.00% |
30.00% |
|||||
II |
No |
1 |
2 |
2 |
5 |
||||
% |
25.00% |
100.00% |
50.00% |
50.00% |
|||||
VI |
No |
0 |
0 |
2 |
2 |
||||
% |
0.00% |
0.00% |
50.00% |
20.00% |
|||||
Total |
No |
4 |
2 |
4 |
10 |
||||
% |
100.00% |
100.00% |
100.00% |
100.00% |
|||||
P = 0.32 |
|||||||||
Table 4 The correlation between different bladder cancer grades and stages & survivin protein expression.
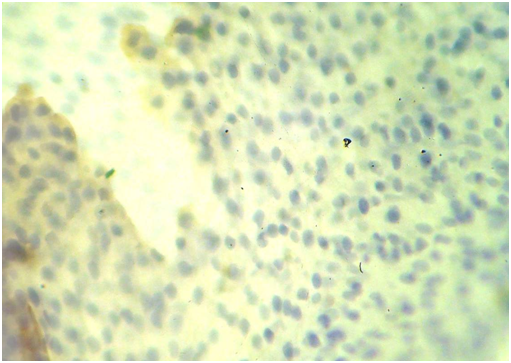
Figure 4 Low grade survivin positive immunostaining some nuclei stained in brown color (blue arrow) due to the over expression of survivin protein inside these nuclei, the other nuclei stained blue (black arrows) as counterstaining (X400).
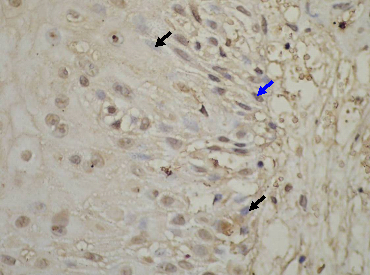
Figure 5 Intermediate grade survivin positive immunostaining many nuclei stained in brown color (blue arrow) due to the over expression of survivin protein inside these nuclei, the other nuclei stained blue (black arrows) as counterstaining (X400).
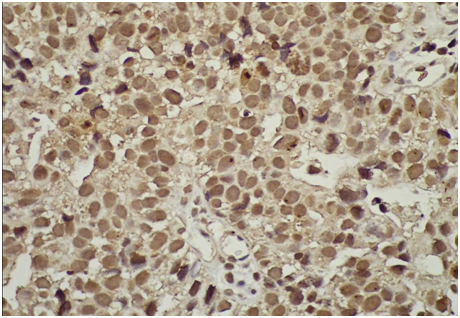
Figure 6 High grade survivin positive immunostaining most nuclei stained in brown color (blue arrow) due to the over expression of survivin protein inside these nuclei, some nuclei show mitosis (X400).
The patient group could be divided into three subgroups according to protein expression levels: law (4 samples), moderate (2 samples) and high (4 samples). On the other hand all samples in the control group show negative IHC surviving staining.
Assessment of the correlation between different bladder cancer grades and stages & survivin protein expression revealed that:
The present investigation was carried out on 26 patients with bladder cancer, the survivin protein expression in paraffin embedded tumor tissues were detected by immunohistochemical staining. Relative expression of survivin mRNA in fresh-frozen tumor tissues was detected by using Taq-Man probe real time PCR analysis. In the present study the ages of the bladder cancer patients ranged from 36–79-years with means±SD 58.96±9.45 and median age 59 years.
This result is closely related to Zapatero et al.20 who reported a median age of 60 years (range 41-77) and Gupta et al.,21 study who found that the mean±SD age was 60.2±4.4 years old (range: 18–90 years old). However, the median age of 63 years (range 42-75) was reported by Zietman et al.,22 and a median age of 63 years (range 31-79) reported by Rodel et al.23
From the present study it was noticed that males were more affected than females, 22 males (84.61%) and 4 females (15.38%) this results is closely related to the previous results of the data reported by Tonoli et al.,24 where males represent 86% of the whole series and Gupta et al.21 who found that the male to female ratio was 8.6:1.21
From the present study it was noticed that the transitional cell carcinoma represented the great majority (22 cases constituting 84.61%), adenocarcinoma (2 cases constituting 7.96%). While small cell carcinoma as well as squamous cell carcinoma each of them constituted 3.85% (1 case per each type). This is was similar to results reported by Lynch et al.3 who stated that most of the tumors 72.0% were transitional cell carcinomas and 22.0% were adenocarcinomas.3
From the present study it was noticed that most of the patients (16 patients, 61.54% ) had tumors with grade III ( 5 patients ,19.23 %) had tumors with grade II, and (5 patients,19.23%) had grade I, However our data was close to Rodel et al.23 where high grade tumors represented 57.8% only,23 However, that was different from Zietman et al.20 who found that high grade tumors represent 77.7%, and also different from Zapatero et al.,22 who reported that 94% of the tumors were of high grade.20,22
The present study showed that 9 patients (34.61 %) had T3 disease, while T2 disease was seen in 8 patients (30.77%), 7patients (26.9%) had T4 disease and 2 patients (7.69%) with T1 disease stage, this result is consistent with the disease stage reported by Shariat et al.25 who noticed that most of the patients (36,5%) had T3 disease, but different in the percent of other patient disease stages. However, this was different from the disease stage reported by Kaufman et al.26 where T2 disease constituted 88% and T3 disease constituted 12% of the entire patient population.26
The size of the bladder tumors among patients of the present study ranged from 1.5-12 cm with mean±SD 6.67 cm±2.6. half of the patients (50%) had tumors 5 - 10 cm in size, 38.5% had tumors 1- 5 cm in size, and 11.5 % had tumors>10cm in size, on the other hand sabaa et al.,27 reported that most of the patients(73.01%) had tumors less than 3cm in size.27 In the present result it was noticed that there was 19 patients (73.07%) without nodal metastasis only 7 patients (26.9%) had variable degree of nodal metastasis.
From the present study it was noticed that there is a positive significant correlation between survivin mRNA expression levels and tumor grade progression as follows: patients had GI disease had mean±SD expression level of 64.5±54.9.Patients with GII disease had mean±SD expression level of 461.64±975.65 and patients with GIII disease had mean±SD expression level of 1684.4±1307.67 with p value of 0.021. So there is a positive significant correlation between tumor grade progression and survivin expression level, so survivin expression increase disease progression. Higher levels of expression were associated with a reduced time to recurrence in noninvasive TCCs (p=0.027). On the other hand, Srivastava et al.28 noticed that survivin concentration did not show any association (p=0.349) with grade.28
According to the present study patients with early stage disease (stage I,II) had mean±SD survivin mRNA expression level of 802.08±652.5 and patients with late stage disease (stage III,IV) had mean±SD survivin mRNA expression level of 1403.78±865,the p value=0.04 , so there is a significant positive correlation between survivin expression and disease stage progression, from this result we can conclude that increasing survivin expression may play a role in the progression of bladder cancer. This result is in consistent with the research results of Srivastava et al.28 noticed that survivin concentration showed direct and significant (p<0.001) correlation with stage, also our results is in agreement with the results of Weikert et al.29 who noticed that Survivin transcripts were commonly detected in tumor tissues, but not in normal urothelium, and increasing mRNA levels correlate with progressing pathologic stage (p=0.001) and grade categories (p<0.004). While the study of Shariat et al.18 showed a highly significant association of Survivin with muscle invasion as there study contain 726 patients in a multiinstitutional study.
The present study also showed that patients group without nodal metastasis (N0) had mean±SD survivin mRNA expression level of 894.8±1129.7, patients group with (N1) nodal metastasis had mean±SD survivin mRNA expression level of 1806.6±772 and patients group with (N2) nodal metastasis had mean±SD survivin mRNA expression level of 2141±1642.3, the p value=0.176, so there was no significant correlation between nodal metastasis and survivin expression in our patients group. On the other hand Shariat et al.,18 noticed that survivin was expressed in 41 of 50 (82%) of the radical cystectomy specimens and 47 of 50 (94%) of the matched lymph node specimens., also they reported that the proportion of specimens with survivin expression increased from advanced TCC (64%) to cancerous lymph node tissue (94%). In patients who underwent radical cystectomy, survivin over-expression was significantly associated with advanced pathologic stage, metastases to lymph nodes, lympho-vascular invasion, number positive lymph nodes, percent positive lymph nodes, disease recurrence, disease progression, and bladder cancer-specific mortality, also Srivastava et al. noticed that survivin concentration showed direct and significant (p<0.001) correlation with nodal status and cytology.
According to the present study patients suffering from adenocarcinoma had mean±SD survivin mRNA expression level of 384±181.02, patients suffering from SCC had mean survivin mRNA expression level of 9.18 without SD (one patient only), patients suffering from small cell carcinoma had mean survivin mRNA expression level of 90.5 without SD (one patient only) and patients suffering from TCC had mean±SD survivin mRNA expression level of 1346.06±1322.8, the p value of this relation was 0.465, so there was no significant correlation between pathological finding and survivin expression in our patients group.
According also to this study, immunohistochemical staining detecting survivin protein expression from 10 paraffin embedded tissues showed that; 4 patients had weak expression level (40%), 2 had moderate level (20%) while the remaining 4 samples showed high expression level (40%). On the other hand all samples in control group showed negative IHC survivin staining.
According to the level of protein expression by immunohistochemical staining, 4 patients with weak staining (100%)had grade 1 disease, 2 patients with moderate level( 100%) had grade 2 and the all 4 with high level of protein expression (100%) had grade 3, indicating that there is a positive correlation between survivin protein expression level and tumor grades p=0.0001.
According also to protein expression; 3 patients with weak staining (75%) had stage1 while the remaining one (25%) had stage 2. The 2 patients with moderate protein expression level (100%) had stage 2. Finally 2out of 4 patients with high staining level (50%) had stage 2 while the remaining 2 in this group had stage 3, indicating that there is no correlation between surviving gene protein expression and different tumor stages. p=0.32. this is somehow different from Weikert et al.29 that showed from the quantitative data that survivin expression increases with both the stage and grade of bladder TCC. They demonstrated a higher expression level in invasive superficial TCC (pT1) than in the noninvasive type (pTa). These findings suggest that survivin may influence the malignant potential of transformed transitional cells and may thus play a role in the progression of bladder cancer.30
In the original report by Swana et al.31 (where expression of survivin was determined by immunohistochemistry, no detectable survivin was reported in normal transitional cells, but it was present in 78% of tumoral specimens.
Further molecular studies should be addressed to elucidate the relation of survivin gene expression and mechanisms of apoptosis and cell division, especially in bladder cancer, as well as that of other molecular markers may help to diagnose patients most likely to benefit from the mode of treatment. Also, from this study we do believe that detection of survivin mRNA by real time PCR is more important than immunohistochemical studies.
None.
The author declares no conflict of interest.

©2017 Arafat, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.