eISSN: 2469-2794


Case Report Volume 2 Issue 3
1Wildlife Institute of India, India
2Survey of India, India
3Amity Institute of Wildlife Sciences, Amity University, India
4Veer Kunwar Singh Universities, India
Correspondence: Surendra Prakash Goyal, Wildlife Forensic and Conservation Genetic Cell, Wildlife Institute of India, Post Box# 18, Chandrabani, Dehradun 248001, Uttarakhand, India, Tel 91-135-2640112-117, Fax 91-135-2640115
Received: June 07, 2016 | Published: June 16, 2016
Citation: Kumar VP, Rajpoot A, Mukesh, et al. Genetic based species identification and tracking of the geographic origin of a fully tanned animal skin in wildlife forensics. Forensic Res Criminol Int J. 2016;2(3):128-134. DOI: 10.15406/frcij.2016.02.00058
Species identification and assign the geographic origin from processed products based on morphological traits is a challenging task in wildlife forensics due to lack of the reference specimens. Here, we report species identification and assign the geographical origin of a fully tanned animal skin using complete mtDNA cytochrome b gene. A nucleotide sequence of 1140 bp cytochrome b was generated from the DNA extracted from the small piece of skin from the inner ear (IEP-01). GenBank BLAST search of the unknown Cyt b (1140bp) sequence against the full range of published Rangifer tarandus has facilitated in identification of species and ascertaining the species of origin with high meta probability (100%). We determined that the seized unknown skin is wild reindeer (R.t. groenlandicus) and has been originated from Canada, where this Least Concern species under the IUCN red list. We propose to establish genetic database across the range of the species threatened due to illegal trade to determine hotspots of poaching.
Keywords: reindeer (rangifer tarandus), forensic genetics, tanned skin, species identification, geographic origin, cytochrome b gene
Illegal trading of wild animals, their body parts and derived products is a major concern in wildlife conservation. Species identification based on DNA analysis is a critical tool in wildlife forensics, especially when the specific morphological characteristics are lost due to transformation or processing.1 Mitochondrial markers, such as cytochrome b, 16S rRNA, 12S rRNA and Cytochrome Oxidase showing interspecific variation, are commonly used to perform species identification and phylogenetic studies in wildlife forensics.2‒5 Cyt b is highly informative in mammals and a large database for this genetic marker is already available for different species and subspecies in different geographical ranges, making it especially useful in wildlife forensics.6 Assignment of samples, we need to amplify large fragment of mtDNA gene that show more number of SNPs over a geographic range.
In most of the wildlife forensic cases, the samples received are in highly degraded condition or highly processed, viz., burn bones/bone piece, cooked meat, tanned skin, claws, canines, etc. Hence, the genomic DNA will be in low quantity and quality as well. Therefore, extraction of a good quality DNA from such type of degraded and tanned sample is a challenging task for the wildlife forensic experts. In the present study, we aimed to select proper part to high yield DNA from highly tanned animal skin and amplify the large fragment of mitochondrial gene to identification of species and to ascertain its geographical origin that was seized by the Custom Authority at Indira Gandhi International Airport, New Delhi India.
A highly tanned animal skin was seized by the Custom Officer at Indira Gandhi International Airport, New Delhi, India, during December, 2012 and sent to the Wildlife Institute of India, Dehradun for species identification (Figure 1). The skin at visual inspection appeared to be of a deer species but the morphological hair patterns did not show definite species assignment due to the lack of an extensive collection of reference samples for comparison. A DNA based species identification strategy was devised to overcome this problem.
DNA extraction
Since the sample was highly tanned, we attempted DNA extraction with commercially available Qiagen DN easy Tissue Kit (QIAGEN, Germany) following manufacturer instructions. Three samples from different parts of unknown tanned skin (IEP-1, OSP-2 and OEP-3) were taken for DNA extraction. The tanning of the skin potentially caused DNA degradation, therefore a section of the inner part of the animal’s ear (IEP-1) that seemed to be relatively less affected by the chemical process was selected for DNA extraction and we processed remaining samples i.e. the outer part of skin and ear (OSP-2 and OEP-3) in order to compare the efficiency of DNA extraction from the different parts of the tanned skin. To determine the quality and concentration of DNA obtained, the samples were subjected to gel electrophoresis on a 0.8% agarose gel in 1X TAE buffer and DNA quantified with a UV spectrophotometer (Amersham Pharmacia) (Figure 2A).
PCR Amplification and Sequencing
DNA template were subjected to polymerase chain reaction using universal of Cyt b gene of different size 200bp, 350bp and 1140bp.7‒9 All PCR reactions were carried out on an Applied Biosystems® 2720 Thermal Cycler (ABI) in a total reaction volume of 25 µl containing 10 µl 2X PCR mix buffer (Amresco); 10 µM of each primer, and 4 µl of total DNA. The Thermal cycling consisted of conditions of denaturation step at 94°C for 3 min, 35 cycles of denaturation(94°C for 30 s),annealing (53°C for 45 s) and primer extension (72°C for 40 s) and a final extension step of 10 min for 72°C 10 min. A small volume of PCR products (5 μl) were subjected to electrophoresis on 2% agarose gel and visualized over an UV transilluminator. Extraction and PCR blanks were incorporated into the analysis. One extraction blank was incorporated with three samples extracted, and one PCR blank was subsequently incorporated with every three extracts that were amplified.
In this study, we used only (IEP-1) PCR amplicon for DNA sequencing. Amplified 1140 bp PCR product was purified using Exo-SAP to remove residual oligonucleotides and dNTPs prior to sequencing reaction. The forward and reverse primers were used independently for the sequencing reactions using the Big Dye® Terminator v3.1 Cycle Sequencing kit to generate sequence from both ends. The products were purified using a standard ethanol precipitation method and sequenced on an ABI 3130 Genetic Analyzer (Applied Biosystems, USA).
Data analysis
Cyt b sequence (IEP-1) was cleaned and validated using SEQUENCHER 4.8 (Gene Codes Corporation, Ann Arbor, MI). Multiple sequence alignments were performed using the CLUSTAL W algorithm implemented in BIOEDIT version 7.0.5.3\9.10 The sequence obtained from the unknown skin specimen was compared with the sequences publicly available at GenBank using BLAST search tool of NCBI (http://blast.ncbi.nlm.nih.gov/). All the sequences that showed similarity with the unknown sequence (IEP-1) were downloaded (Tables 1 & 2) and used for phylogenetic analysis using Kimura 2 parameter distance matrix with the neighbor-joining method as implemented in Mega v5.0 software.11
|
Species/subspecies |
Common name |
Origin |
Accessions number |
|
Cervus albirostris |
Thorold’s deer |
China, Qinghai |
AY044863.1 |
|
Cervus nippon |
Sika deer |
China |
AB021093.1 |
|
Cervus elaphus |
Red deer |
Germany, Encloser |
AY118198 |
|
Cervus elaphus hippelaphus |
Middle European red deer |
Yugoslavia |
AY070225.1 |
|
Cervus elaphus brauneri |
Krim red deer |
Ukraine |
AY148966.1 |
|
Cervus elaphus hippelaphus |
Middle European red deer |
France |
AY244491.1 |
|
Cervus elaphus scoticus |
Scottish red deer |
Scotland |
AB021099.1 |
|
Cervus elaphus atlanticus |
Red deer |
Norway, Hitra |
AY070226.1 |
|
Cervus elaphus xanthopygus |
Isubra |
Russia, Anjui |
AY070224.1 |
|
Cervus elaphus canadensis |
American wapiti |
North America |
AF423198.1 |
|
Cervus elaphus sibericus |
Siberian wapiti |
China, Mongolia |
AY044862.1 |
|
Cervus elaphus kansuensis |
Kansu red deer |
China, Dong Da Shan |
AY070223.1 |
|
Cervus elaphus macneilli |
MacNeill’s deer |
China, Qinghai |
AY035875.1 |
|
Cervus elaphus wallichi |
Shou |
China, Tibet |
AY044861.1 |
|
Cervus elaphus barbarus |
Barbary red deer |
Tunisia, Tunis |
AY070222.1 |
|
Cervus elaphus corsicanus |
Sardinian deer |
Sardinia |
AY244489.1 |
|
Cervus elaphus maral |
Maral |
Iran |
AF489280.1 |
|
Cervus elaphus songaricus |
Tien Shan wapiti |
China, Tien Shan- |
AY035871.1 |
|
Cervus elaphus hispanicus |
Spanish red deer |
Spain, La Gaganta |
AF489281.1 |
|
Cervus elaphus hippelaphus |
Middle European deer |
Bulgaria |
AF423195.1 |
|
Cervus elaphus bactrianus |
Bactrian red deer |
Tadzikistan |
AY142327.1 |
|
Cervus elaphus yarkandensis |
Yarkand red deer |
China |
AY142326.1 |
|
Cervus unicolor |
Sambar |
India |
JN861032.1 |
|
Axis |
Chital |
India |
JN596156.1 |
|
Cervus duvaucelii |
Swamp deer |
India |
EF079830.1 |
|
Axis porcinus |
Hog deer |
Germany |
AY035874.1 |
|
Cervus eldi thamin |
Thamin deer |
Thailand |
EF079829.1 |
|
Rangifer tarandus |
Reindeer |
Norway |
DQ673123.1 |
Table 1 List of GenBank accession numbers used in this study and their respective names with geographic origin
|
Haplotype |
Species Name |
Wild/ Semi domestic |
Geographical Origin |
Accessions number |
|
Hap-01 |
R. tarandus |
Semi domestic |
Norway |
DQ673123.1 |
|
Hap-02,03, 04,05 |
R. tarandus |
Semi domestic |
Alaska/Russia, |
DQ673122.1,DQ673127.1, DQ673130.1,DQ673131.1 |
|
Hap-06,07 |
R. tarandus |
Semi domestic |
Svalbard Is, Sweden |
DQ673124.1,DQ673125.1 |
|
Hap-08,09, 10,11 |
R. tarandus |
Semi domestic |
Russia |
DQ673126.1,DQ673132.1, DQ673133.1,DQ673135.1 |
|
Hap-12,13,14 |
R. tarandus |
Semi domestic |
Alaska |
DQ673128.1,DQ673129.1, DQ673134.1 AY726672.1,AY726673.1, |
|
Hap-15,16,17,18, 19,20 |
R.t.caribou |
Wild |
Canada |
AY726674.1,AY726677.1, AY726675.1,AY726676.1 AY726679.1,AY726691.1, |
|
Hap-21,22,23,24,25,26 |
R.t.groenlandicus |
Wild |
Canada |
AY726705.1,AY726706.1, AY726720.1,AY726729.1 |
|
Hap-27,28,29,30,31,32, 33,34,35,36,37,38,39,40,41,42,43,44,45, 46,47,48,49,50,51,52,53,54,55,56, 57,58,59,60,61,62,63,64,65,66, 67,68,69,70 |
R.t.granti |
Wild |
USA |
AY726680.1,AY726682.1, AY726685.1,AY726690.1, AY726697.1,AY726701.1, AY726724.1,AY726730.1, AY726703.1,AY726711.1, AY726712.1,AY726714.1, AY726715.1,AY726718.1, AY726721.1,AY726723.1, AY726728.1,AY726683.1, AY726686.1, AY72689.1, AY726700.1,AY726702.1, AY726684.1,AY726704.1, AY726707.1,AY726719.1, AY726720.1,AY726726.1, AY726687.1,AY726688.1, AY726693.1,AY726696.1, AY726699.1,AY726692.1, AY726695.1,AY726713.1, AY726722.1,AY726694.1, AY726698.1,AY726709.1, AY726717.1 |
Of the total samples (n=3), DNA extracted from, two samples (OSP-2 and OEP-3) did not yield good DNA (Figure 2A) and amplification was not observed after PCR (Figures 2A & 2B). Sample IEP-1 yielded detectable product of all three different size (200bp, 350bp and 1140bp) Cyt b gene on agarose gel electrophoresis (Figure 2B).
In case of non amplified samples (OSP-2 and OEP-3) we suspected the presence of PCR inhibitors within the DNA, or inhibitors may be due to chemical used in tanning process or from the environment. Therefore, we tested the DNA for PCR inhibitors, by using known positive PCR reactions. The results indicated that two DNA samples (OSP-2 and OEP-3) contained PCR inhibitors. To solved this problem, we check various dilution (1x-100x) of DNA sample, we found that 50x diluted DNA give PCR amplification in short fragment of Cyt b gene (200 bp) but remaining fragments of Cyt b gene were not amplified (Figure 2C).

Figure 2 Agarose gel electrophoresis of unknown animal skin sample. (A). Quality of Genomic DNA, Lane 1,2,3 and 4 represent IEP-1, OSP-2, OEP-3 and –Ve control respectively. (B) Amplified PCR product, Lane 2, 3, 4 and 5 indicate IEP-1, OSP-2, OEP-3 and –Ve control respectively. Only Unknown skin sample IEP-1 show amplification in all three different sized primer (200bp, 350bp and 1140bp). (C) Amplification after 50x dilution Lane 2 to 3 is of OSP-2 and OEP-3 respectively. Indicate amplification in 200bp only and remaining primer did not amplify. Lane 4 and 5 are showed +Ve and -Ve control. Lane 1 in (B) and (C) show 100 bp DNA ladder.
Tanning is an aggressive chemical process that potentially damage DNA and this was the probable cause for low yield of DNA in extraction process from the outer parts of the skin sample, which were visibly fully tanned. UV radiation and overexposing to heat are also known to cause DNA damages12,13 and may have contributed to the negative amplification. The inner ear fragment was probably less damaged by tanning chemicals, UV radiation and heat and resulted in yielding good quality PCR product. BLAST analysis of Cyt b gene indicated that the samples of unknown tanned animal skin (IEP-1) showed 100% similarity Rangifer tarandus.
The neighbor-joining phylogenetic tree with other deer subspecies also showed that the unknown skin (IEP-1) is belonged to the Rangifer tarandus species, commonly known as reindeer, with a strong bootstrap value of 100% (Figure 3). However, five subspecies of wild reindeer are currently recognized: Barren-ground caribou (R. t. groenlandicus), woodland caribou (R. t. caribou), Grant’s caribou (R. t. granti), Peary caribou (R. t. pearyi), Dawson’s caribou (R. t. dawsoni, extinct), and are still widely distributed across northern Eurasia and North America (caribou).13,14 Today, almost 50% of the approximate 3,00,0000 reindeer in the Old World are wild animals, and some of this population are semi-domestic (R. t. tarandus) which are managed in close coexistence in many areas.15,16 We compared complete sequence of cytochrome b (1140 bp) gene of unknown tanned skin sample (IEP-1) with three subspecies of wild reindeer and semi-domestic reindeer sequence taken from Cronin et al.,16,17 available in GenBank (Table 2). The Multiple Sequence Alignments displayed 100% similarity with wild reindeer Barren-ground caribou (Table 3).
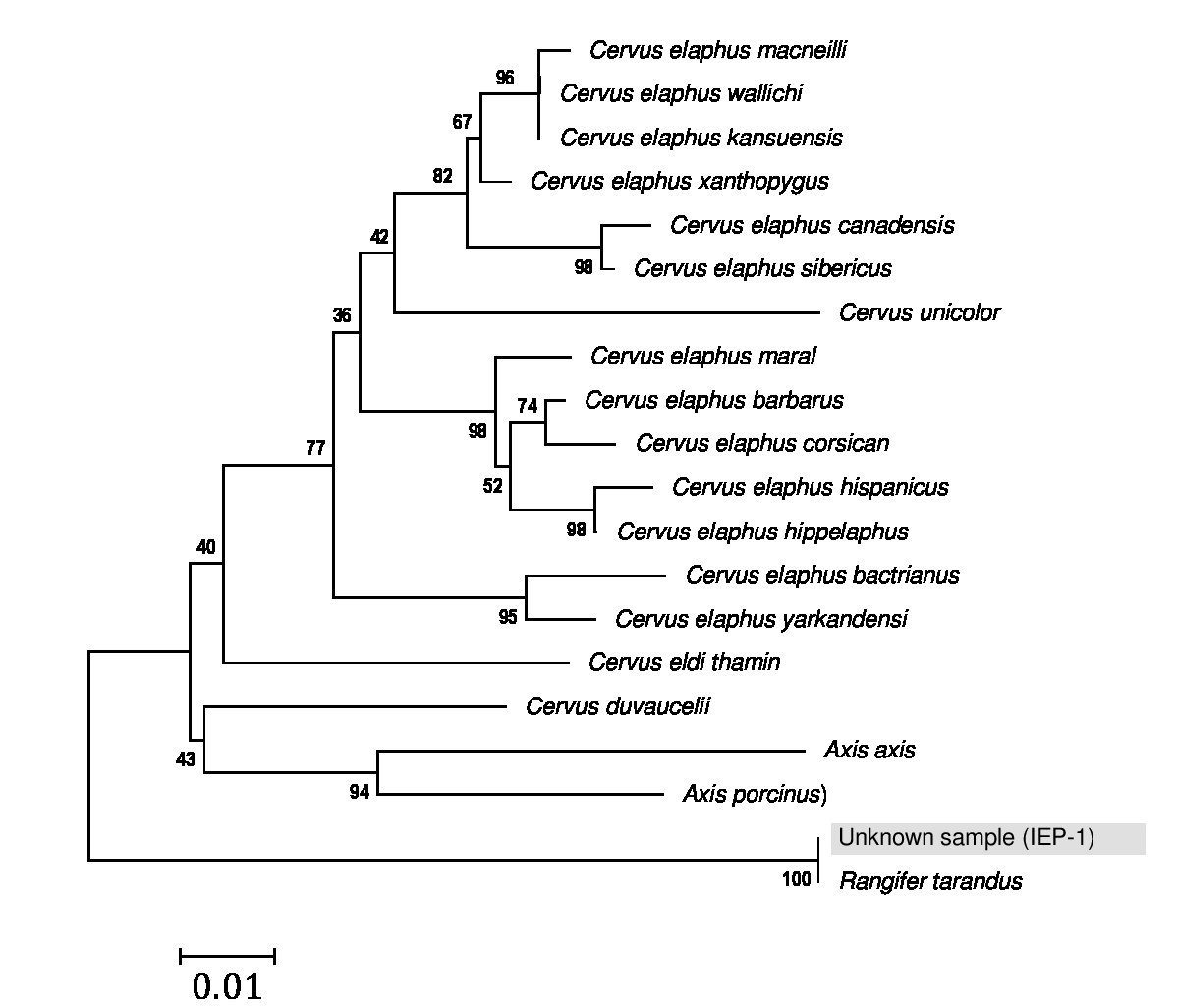
Figure 3 Neighbor-joining phylogenetic tree showing the relationships of unknown skin sample with the other deer species of the world.
|
Unknown sample |
Subspecies with the highest similarity (genbank accession) |
Query coverage (%) |
Similarity (%) |
Hap/geographic origin |
|
IEP-1 |
Rangifer tarandus |
100 |
99 |
Hap01/Norway |
|
IEP-1 |
Rangifer tarandus |
100 |
99 |
Hap-02,03.04,05 / All from Alaska/Russia |
|
IEP-1 |
Rangifer tarandus |
100 |
99 |
Hap-06 / Svalbard Is |
|
IEP-1 |
Rangifer tarandus |
100 |
99 |
Hap-07 / Swden |
|
IEP-1 |
Rangifer tarandus |
100 |
99 |
Hap-08,09,10,11 All from Russia |
|
IEP-1 |
Rangifer tarandus |
100 |
99 |
Hap-12,13,14 / All from Alaska |
|
IEP-1 |
Rangifer tarandus groenlandicus |
100 |
100 |
Hap-21,22,23,24,25,26/ Canada |
|
IEP-1 |
Rangifer tarandus granti |
100 |
99 |
Hap-22, 24,27/ USA |
|
IEP-1 |
Rangifer tarandus caribou |
99 |
98 |
Hap-15,16,17/ all from Canada |
Table 3 Similarities in the Cyt b locus between unknown sample (IEP-1) and sequences of reindeer subspecies available in GenBank
In order to assess the geographic origin of the unknown sample (IEP-1), we retrieved the n=76 sequences of cytochrome b gene (from NCBI Gene Bank) and found 70 haplotypes in these sequences that represent 56 haplotypes from 3 subspecies of wild reindeer; Grant’s caribou, Canadian barren-ground caribou and woodland caribou and 14 haplotypes from different geographic origins of semi-domestic reindeer that were found in 13 herds from 3 regions: Alaska, Russia, and Scandinavia (Table 2).
The Alaskan herds represent the geographic range from Siberia, Russia and extended to the Seward Peninsula, Alaska. While the reindeer from the Russian herds represent geographical range restricted to Magadan, district in Siberia and the Scandinavian herds reindeer are represent the Norway population. The sequence of unknown sample (IEP-1) matched with the Hap-21, 22 wild reindeer that represents the Canadian population with 57% bootstrap value and displayed 100% similarity. The remaining reindeer haplotypes have sequences similarity and bootstrap values which were lower, when compared with unknown sample (IEP-1) (Table 3 & Figure 4).

Figure 4 Haplotype Neighbor-joining phylogenetic tree with 70 different geographically originated showing the relationships of unknown skin sample (IEP-1) with the semi domestic (Hap-01 to Hap-14) and three wild subspecies (Hap-15 to Hap-70) of Reindeer. Details of haplotype given in Table 2.
The unknown query sequence showed 100% similarity with the haplotype H-21, 22 that has been originated from Canada. Therefore, based on the data of Cronin et al.,16,17 we concluded that the animal skin seized by Indian Customs is of the wild reindeer (R. t. groenlandicus) and has been originated from Canada. Wild population of reindeer is Least Concern species in IUCN, listed on Appendix III of the Bern Convention.
In conclusion, we describe the utility of large fragment of cytochrome b (1140 bp) mtDNA gene sequence in the species identification and ascertaining the geographic origin of a fully tanned animal skin. In order to assign the geographical origin of species, it is necessary to use Cyt b gene, which cover the entire range of probable haplotypes. In many study mtDNA Cyt b gene sequences were used to determine and investigate the geographical origin of species.18,19 This result shows the importance of genetic analysis in wildlife forensics and the utility of DNA based analysis in the implementation CITES (Convention of International Trade in Endangered Species). We suggest that DNA analysis of tanned skins should target samples from inner parts of the skins.
Furthermore, we defended that database of Cyt b gene sequences across the range of species threatened due to illegal poaching and classified in Appendix I should be established to allow for confident genetic identifications. Wild reindeer is the least concern species under the IUCN red list. This means that we have a special responsibility to take care of and manage the reindeer in a way that will allow future generations to experience viable population of reindeer. This database would be of great aid in DNA-based investigations of illegal trade, the implementation CITES and the detection of geographic poaching hotspots.
The authors are thankful to the Director, Dean and Research Coordinator, Wildlife Institute of India (WII), Dehradun, for their strong support. We would like to thank Nodal Officer of Wildlife Forensic and Conservation Genetic Cell (WFCGC). We would like to thank all the researchers and staff of the Wildlife Forensic and Conservation Genetic Cell (WFCGC), for providing scientific and technical assistance while undertaking the work.
The author declares that there are no conflicts of interest.

©2016 Kumar, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.