Advances in
eISSN: 2572-8490


Review Article Volume 3 Issue 3
1School of Medicine, Keele University, UK
2Department of Chemistry, Loughborough University, UK
Correspondence: Paul Roach, Department of Chemistry, School of Science, Loughborough University, Leicestershire, LE11 3TU, UK
Received: November 08, 2017 | Published: December 15, 2017
Citation: Köse-Dunn M, Fricker RA, Roach P. Tissue engineered organoids for neural network modelling. Adv Tissue Eng Regen Med Open Access. 2017;3(3):391–401. DOI: 10.15406/atroa.2017.03.00066
The increased prevalence of neurological diseases across the world has stimulated a great deal of research into the physiological and pathological brain, both at clinical and pre-clinical level. This has led to the development of many sophisticated tissue engineered neural models, presenting greater cellular complexity to better mimic the central nervous system niche environment. These have been developed with the ambition to improve pre-clinical assessment of pharma and cellular therapies, as well as better understand this tissue type and its function/dysfunction. This review covers the necessary considerations in in vitro model design, along with recent advances in 2Dculture systems, to 3D organoids and bio-artificial organs.
Keywords: neural, neuronal, organoid, tissue engineering, pre-clinical
CNS, central nervous system; iPSC, induced pluripotent stem cells; HD, huntington’s disease; PD, parkinson’s disease; AD, alzheimer’s disease; ALS, amyotrophic lateral sclerosis; MS, multiple sclerosis; TBI, traumatic brain injury; NSC, neural stem cells; PDMS, poly(dimethylsiloxane)
The use of cells as building materials provides a powerful tool to the fields of both regenerative medicine as a broad aspect and in particular to tissue engineering, with the potential to deliver a tremendous amount of information both in vivo as cell-based therapies and/or in vitro as cell models. Combining cells with specialized biomaterials, suitable biochemical growth/differentiation factors, extracellular matrices (‘scaffolds’) and diverse biomimetic environments creates a myriad of opportunities for extensive study of tissues in both physiological and pathological forms, and the creation of strategies for regenerating damaged tissues.1,2 Due to the complexity of living tissue, with multiple cell types acting in synergy to give the whole tissue its function, there are many efforts to model tissues in vitro. For the most part such modelling aims strikes a balance between the ability to create functional tissue structures and the simplification in the complexity observed in vivo. Organoids have been highlighted as one of the major advances in developing suitable models for various specific tissue types. Amongst these are intestine3 lung4–6 and kidney,7,8 to name a few. Further models of heart, cartilage and skin, as well as functional systems such as the vascular, endocrine, musculoskeletal, and nervous systems have been reviewed by Benam et al.9 Body- and human-on-a-chip systems further aim to draw connectivity between each of these separate models in order to mimic basic physiological function on a larger scale.10,11
Tissue models must present a reasonable mimic of normal physiological function in order that they are deemed useful; it is this ‘use’ which is now expanding as we gradually increase our micro/nanofabrication capabilities to guide complex tissue engineering approaches, better replicating normal and diseased function. Our models allow for better understanding of function and dysfunction, disease spread and how efforts for treatment may be optimised. Pre clinical assessment of disease is certainly one of the tissue engineering ‘holy grails’ with personalised medicine approaches being a major future ambition for research within this area. In this review we cover those developed for the central nervous system (CNS), namely neural tissue engineering, which remain one of the most challenging tissue engineering areas due to the complexity of interconnectivity and communication between the plethora of cell types, all requiring very specific architecture on the cell-level to infer function. In this review we include the range of approaches used for both normal and diseased CNS models.
Neural tissue engineering reaches a new level of complexity due to the fact that unlike other tissues, the structure and distinct architecture is seen even at the cellular level and is vital to assure functionality. The human brain contains many combinations of intricate micro- and macroscopic12 connections- whether morphological, functional, or both-occurring at specific spatial nodes at specific temporal intervals, creating an extremely complex network between vast cell populations, making the brain extremely challenging to model in vitro. An average adult human brain will have a mass of 1.5 kg containing100 billion neuronal cells (neurons, which transmit and receive information through electrical signals, forming synapses with other neurons) and 1 trillion non-neuronal cells (glia, structural cells of the brain, composed mainly of astrocytes, oligodendrocytes and microglia).13 In addition, each neuron can connect with other neurons (and astrocytes14) via synapses, at an average of 7000 connections per neuron, resulting in approx. 100 trillion connections in the adult human brain.15,16
Due to this complexity, it has been difficult to develop living artificial neural networks that are reproducible, can be scaled-up and are low-cost, reliable, as well as efficient, robust, and reproducible,1,2 both during standard physiological situations and abnormal pathological situations during disease.17,18 Some of the many design considerations for neural models are highlighted in Table 1.
Design consideration |
Culture options |
Brain area to model |
Most common areas reported in literature are cortex, hippocampus, cerebellum, spinal cord and sensory ganglia. |
Cell culture class |
Primary cells and secondary cell lines. Primary cells are isolated directly from |
Cell developmental age |
As the brain develops cell types mature into the various populations that may be required. |
Similarities to the human nervous system |
Can use adult human iPS cells, embryonic/foetal human cells, animal cell lines or |
Culture type |
Can culture tissue/organ slices or dissected disperse individual cell populations. |
Preservation of in vivo organisation |
Neural cells can grow effectively in a 2D planar culture (monolayer) or 3D matrix |
Electrophysiological integrity |
Are the cells functional within the model? The neural cells within the model should have |
Culture environment |
Extracellular fluid composition, temperature, pH, gas phase, substrate, dimensions. |
Model material |
Non-toxic, non-inflammatory, non-allergenic, non-carcinogenic, light, soft, mechanically durable and chemically stable |
Table 1 What to consider when planning an in vitro neural model, adapted from19
There are multiple ways to categorise neurological diseases, but the major types include those caused by: genetic disorders (Huntington’s disease (HD)20 and muscular dystrophy21); cellular degeneration (Parkinson’s disease (PD)22 and Alzheimer’s disease (AD)23); movement disorders (neuromuscular disease such as amyotrophic lateral sclerosis (ALS)24 and multiple sclerosis (MS)25); damage to central nervous system blood supply (stroke26); electrophysiological disorders (epilepsy27); physical injury (spinal cord injury (SCI),28 traumatic brain injury (TBI)29); cancer (such as glial and non-glial tumours30) and infection (bacterial meningitis31).
The drivers for the development of tissue engineered models of disease revolve around our current inability to understand (dys) function of the CNS and further how to better treat neurodegenerative disorders that are becoming increasing prevalent. Large pharmaceutical companies have spent billions of dollars over the past decade trying to address these issues, but have now stepped back from major funding efforts due to lack of progression. With increasing demand to new therapies, both pharmaceutical and cellular therapies, and our increasing capabilities to better fabricate tissues with a degree of complexity, tissue engineered models are providing a stepping stone for the research of new ways to move this research area forward. Key features of the diseases and disorders being of high interest by this growing sub-field of regenerative medicine are highlighted in Table 2.
Stroke (traumatic) |
Epilepsy |
Parkinson’s disease |
Huntington’s disease |
Alzheimer’s disease |
Amyotrophic lateral sclerosis |
|
Prevalence |
Second most common form of death, 16 million |
Most common neurological disorder, 65 |
5.70 per 100,000 in Europe, North America and Australia.20 |
Approx. 0.4% of world pop. (26.6 million), |
2.2 per 100,000 per year in Europe, with peak |
|
Main Symptoms |
Unilateral inability to move and loss of |
Seizures: from momentary loss of awareness to |
Cognitive and behavioural limitations resulting |
Chorea, general physical instability, cognitive |
Progressively worsen with age, from mild |
Cognitive function is largely unaffected in |
Causes |
30-40% cases idiopathic, of the remainder 87% |
60% cases idiopathic. Other cases result after |
Most cases are idiopathic, a minority of cases |
Autosomal-dominant disorder caused by a |
Mostly idiopathic, can be familial with 49-79%
|
5-10% of cases inherited, potentially due to |
Pathology |
Irregular levels of blood in the brain |
Electrophysiological dysfunction in the brain, |
Loss of dopaminergic neurons in the substantia |
Huntingtingene codes for huntingtin protein in |
Degeneration of temporal and parietal lobes, |
Death of upper and lower motor neurons in the |
Table 2 Comparison of the key components of the major neurological disorders
The vast majority of neurological diseases are not well understood. This lack of knowledge concerning causative mechanisms of human neurodegenerative disease outlines the vital importance of developing efficient pre-clinical research methods, especially when taking into account the prevalence and fatality of some of the diseases. More effective pre-clinical assessment is needed to address the fundamental underlying mechanisms behind develop in neurological disorders, so as to further refine how we model diseases in pre-clinical research. Current pre-clinical models are unable to accurately predict the efficacy of pharmaceuticals or cellular therapies within human disease patients. Dr Don Ingber, Director of Harvard University’s Wyss Institute of Biologically Inspired Engineering (July/August 2012, MIT Tech. Review) has spoken out about this in a topical interview:
“… the drug development model is actually broken. It takes many, many years to get a drug to market, it’s incredibly expensive, innumerable animal lives are lost – and then the results from animals ‘usually’ don’t predict what happens in human. So this is a huge cost to the economy and to the pharmaceutical industry.”
There is a need for new pre-clinical models to determine at a much earlier stage whether the treatment in question is going to be effective, thus eliminating unnecessary clinical trials. Current pre-clinical models can be either in vivo or in vitro. But there is often a considerable lack of diseased tissue for study, especially concerning human models, due to biopsies involving limited environmental control, variable sample thickness, and destruction of countless input/output connections from both neuronal and glial cells53 upon biopsy. Therefore, pre-clinical work usually relies upon animal models, either as a whole for in vivo study, or as a source of brain slices or neuronal/glial cells for in vitro culture.
Animal models are a necessary regulatory hurdle for any medical therapy, although there are well known pitfalls resulting from the difficulties of measurement or understanding of mode of therapy action in the CNS, or indeed difficulty to measure the output of effect; these are not least beset by the ethical considerations of using animals for research, nor their non-accurate mimics for human assessment. Whole animal models are limited by the behavioural outputs, alongside the treatment zone of either pharmaceutical or cellular therapy delivery encompassing a large volume if not the whole tissue. The same is observed even for tissue slices, albeit that these do offer a larger sampling size from a single animal and potential to better interrogate tissue at the cell level. For in vitro developed models the choice of cells and their presentation are the initial key design factors. Organoid models do permit in some cases real-time monitoring of tissue by microscopy and function by e.g. electrode insertion, although the complexity of these cannot realistically achieve anything like that observed for normal CNS tissue. Here we give some insight into selection considerations made during the development of these in vitro systems.
The most basic in vitro neural models make use of populations of neurons being used after removal or culture of the cells to isolate specifically neurons rather than to include glial cells. The absence of glia markedly decreases the accuracy of the model as a mimic of the in vivo CNS, where astrocytes and other glial cells play a vital role in the brains structure and function. As most biopsies and samples from brains feature a physiologically-relevant mixture of neuronal and glial cells, neuronal-only cell models are often derived from exogenous sources such as neural stem cells (NSCs), artificial sources such as iPSC cells or cancer cell lines such as the human neuroblastoma cell line SH SY5Y, all discussed further below. Despite the progress made with neuronal-only in vitro cell models, the absence of glia is a major disadvantage of these models, only with a physiologically relevant mixture of neuronal and glial cells can we approach a good mimic of the highly complex in vivo CNS niche. Such mixed cultures can be isolated from in vivo sources such as the Sprague-Dawley rat.54 The combination of both makes primary neuronal-glial cultures a powerful in vitro model for studying neurological disease.
Induced pluripotent stem cells
Due to their pluripotent nature iPSCs can differentiate down a neural pathway and be used to generate neural tissues. By using neurologically-affected sources (e.g. obtained from PD patients) iPSCs can be used to model diseases, being genetically identical to the disease source. Table 3 features a list of recent iPSC neurodegenerative disease models: While iPSCs are a versatile tool for studying neurological disease, sometimes referred to as a ‘disease-in-a-dish’ model, they have several limitations. There are only a limited number of individual lines used to model disease mechanisms, and all of them exhibit significant biological variance, making them somewhat unpredictable with decreasing reproducibility between experiments.55,56 Such variability results from the reprogramming process, culture-induced differences due to the lack of robust differentiation protocols, and differences in genetic background between patients.57
Disease type |
Cell modelled |
Results |
Reference |
Parkinson’s |
Dopaminergic neurons |
Fibroblasts from five PD patients were |
|
Parkinson’s |
Dopaminergic neurons |
Fibroblasts from PD patients differentiated |
|
Familial alzheimer’s |
Neurons |
Fibroblasts with presenilin 1/2 mutation (a |
|
Alzheimer’s in down syndrome (trisomy 21) |
Cortical neurons |
Generated cortical neurons developed AD |
|
Alzheimer’s (familial and sporadic) |
Neurons |
Generated neurons showed normal |
|
Alzheimer’s (familial and sporadic) |
Neurons |
Generated neurons showed Aβ oligomer |
|
ALS (familial) |
Motor neurons |
Generated motor neurons formed cytosolic |
|
ALS |
Motor neurons |
Generated motor neurons contained SOD1 |
|
ALS |
Motor neurons |
Generated motor neurons expressed markers of |
Table 3 Recent iPSC models of neurodegenerative diseases
Neural stem cells
There is a growing interest in using stem cells for the treatment of neurodegenerative diseases, especially multipotent stem cells with a neural origin. Several studies have used NSCs as a neuronal-only in vitro neural cell model. NSCs were used as a transplant by Ono et al. during the development of an in vitro NSC model of schizophrenia, using NSCs from E13.5 foetal rats to repair damage to primary cortical neural cultures from E18 foetal rats. NMDA receptor antagonist MK-801 as well as serum/nutrient deprivation stress was used to damage the cortical neurons, and exogenous NSCs were transplanted to determine any neuroprotective effects. This study found that NSCs exerted neuroprotective effects, altered cell survival signalling by indirect cell-cell contact, restoration of protein levels (reduced by the stress) and had a general anti-apoptotic effect on cells affected by both forms of damage, rescuing the damaged cortical neurons.67
Another effect of NSCs on damaged neural cell populations is a paracrine effect via the release of exosomes, affecting cell-cell communication. Bonafede et al.68 developed an in vitro model of ALS through motor neuron-like NSC-34 cells (NSCs that over express human ALS mutations SOD1 (G93A, G37R or A4V)) that exhibited oxidative stress found in ALS in vivo. The NSC-34 cells were protected from this stress, increasing cell viability, by treating the cells with exosomes derived from murine adipose-derived stromal cells. The study promotes exosomes as a potential therapy in motor neuron disease.
NSCs represent a flexible platform through both their ability to differentiate into multiple cell types in vitro, but their ability to be genetically modified to better match the diseases they are modelling (as exampled above with the NSC-34 mutants exhibiting oxidative stress similar to that found in ALS). NSCs have been genetically modified to stably express and secrete neprilysin (known under several names, including neutral endopeptidase or NEP), an enzyme that degrades beta-amyloid protein (Aβ), one of the causative elements of Alzheimer’s disease. In this study, NEP-expressing NSCs were found to significantly reduce Aβ pathology when transplanted, in both proximal and distal areas.69 Further use of NSCs (as well as embryonic stem cells (ESCs) and iPSCs) as in vitro neural models of disease is summarised by.70
SH-SY5Y human neuroblastoma cell line
Primary cells derived from the CNS are limited in that once they differentiate and mature into neurons they reach a static population and propagate no further. The advantages of using a cancer cell line such as the SH-SY5Y neuroblastoma cell line is that they can be cultured indefinitely, and as such the line is often used for in vitro neural models of disease, also due to their availability, ease of culture and exhibition of dopaminergic markers. This cell line has been especially useful for modelling Parkinson’s disease. However, Kovalevich et al.71 identify three characteristics of SH SY5Y cells that should be considered for in vitro studies. Firstly, SH-SY5Y cultures include adherent (cells that grow when attached to surfaces) and floating cells (cells that grow unattached), with the floating cells having a unique phenotype but mostly discarded during media changes, the focus being on the adherents. Secondly, SH-SY5Y cultures produce both neuroblast-like (N) and epithelial-like (S) subtypes, with only the N morphology exhibiting dopaminergic markers and enzymatic activity. However, the N-type cells can be specifically selected for by forcing the SH-SY5Y cells to differentiate to a mature neuron-like phenotype, which is the final characteristic, the most common means of differentiation being treatment with retinoic acid (RA).71–73
Due to their expression of dopaminergic markers, SH-SY5Y cells are used most frequently to model Parkinson’s disease in vitro. A recent review of these studies demonstrates several such models,74 where Parkinson’s disease is simulated in a number of ways. One method involves over expressing α-synuclein (or the A53T/A30P mutants),75,76 to varying success. A more popular method involves mimicking abnormal mitochondrial function and the associated oxidative stress and autophagy with the use of specific drugs such as MPP+,77,78 6-OHDA79,80 and paraquat;81,82 or through gene knockouts such as PINK1 silencing.83
Co-culture models
The interaction between neurons and astrocytes is a vital component to include in any in vitro neural model, as demonstrated by an ALS model where mutant SOD1 motor neurons were adversely affected when cultured with mutant glial cells, with the glia having a direct, non-cell autonomous effect on motor neuron survival.84 Other studies have made use of co-cultures for observation of pharma effects on gene regulation. Nissou et al.85 presented work on vitamin D deficiencies within neuronal/glia co-cultures, highlighting 17 genes related to neurodegenerative diseases, 10 of these encoding proteins potentially limiting the progression of Alzheimer’s disease.
Wang et al.86 have presented models co-culturing neuronal-glia mixtures in similar ratios as would be observed in vivo: 37% neurons, 51% astrocytes, 7% microglia and 5% other cells, after 14days culture in vitro (DIV).These models use the complex physiological neuron-neuron, glia-glia and neuron-glia interactions to increase the accuracy of the model to the in vivo environment, as well as the fact that they can be obtained from various brain regions (cortex, subcortical nuclei, hippocampus, etc) to demonstrate regional differences in susceptibility during certain neurodegenerative diseases, such as Parkinson’s and Huntington’s disease which are mainly localised to the basal ganglia region, and generally how neurons and glia from different regions are inherently different.87
The presence of astrocytes and microglia in culture give these cultures the ability to better model certain diseases, especially diseases where inflammation is involved as an important disease modifying factor, considering that microglia and astrocytes are involved in secretion of inflammatory mediating factors.88 Microglia in particular play an important role in injury and recovery, as shown when activated microglia mediate damage to injured dopaminergic cells, showing how inflammatory reactions could specifically target oxidative injuries.89 As well as being cultured together as a mixture, neurons and glia can also be co-cultured in a segregated manner, with neurons and glia actively signalling each other while not being in contact.90 The activation of glia being significant, as this activation has been shown to play a role in the pathogenesis of various neurodegenerative diseases.91 However, generating segregated cultures demonstrates a contradiction: how can different cell cultures be physically separated from each other while still being allowed to communicate (otherwise the result is simply two isolated cultures), creating a segregated co culture? One effective answer is the use of micro-scale features to allow only the processes (axons, dendrites, generalised as neurites or processes) from each culture to interact, with the cell bodies themselves segregated. To this end, process outgrowth must be directed and controlled, often using chemical patterns or micro-channels.
Micro- and nano-scale physical features have a marked effect on cell culture; cells experience the features through mechanotransduction and undergo biochemical, morphological and genetic alterations.92 For example, growing human NSCs on micro-scale grooves resulted in elongation and bipolar growth, with the cells aligning to the grooves and growing along the groove wall.93 Primary cells have also been used to develop direction cues as would be observed in vivo, using radial glia to guide neurons.94 This ability to align and direct cells allows control over the direction of neuron outgrowth, and the formation of segregated neuronal-glial ‘circuits’. There is a wealth of literature with many reviews on the topics of surface texturing, chemical patterning and cell control.95
The ability to segregate and direct neural cells has evolved over the last 40 years or more, starting with the Campenot chamber in 1977,96,97 which isolated processes of long-projection neurons using a Teflon TM barrier and micro-scale grooves. The chamber was modified to accommodate all types of neurons, including those with shorter processes such as inter-neurons.98 Whilst these models allowed the separation of cell body and elongating neurites, they could only accommodate one cell type, with no consideration of co culturing cells at this time. Based on these shortcomings, a new model was developed: two chambers linked by micro-scale channels, fabricated via photo- and soft-lithography and sealed to a surface, resulting in a microfluidic device for controlled segregated cell culture.99–101 This model has formed the basis for microfluidic cell culture devices, being modified to increase in complexity and therefore in effectiveness as an in vitro neural model (a particular example features seven chambers and glial cell interaction102). All of these models can be seen below in Figure 1.
The basic design of microfluidic models demonstrated in Figure 1C-CF have become a gold standard for in vitro neural models due to their many advantages over other model types: unlimited design opportunities allowing for increasingly complex designs over time, very specific localisation of cells and/or chemicals, higher throughput, can be scaled up, highly sequential/parallel experimentation, extremely small volumes of media/chemicals per experiment (reducing cost), micro-channels allow fluidic isolation between compartments stopping the spread of treatments between chambers, greater control over cell patterning/manipulation, greater control over extracellular and cellular microenvironments, visible to conventional microscopes when made with optically transparent material (e.g. PDMS) and are low-cost disposable devices.103,104

The original design seen in Fig.1C has been widely used for a variety of different co-cultures, with recent examples including: cortical neurons,105,106 cortical-cortical and cortical-thalamic co-culture systems,107,108 hippocampal-glial co culture systems,109,110 cortical neurons co-cultured with genetically modified astrocytes,111 embryonic forebrain neurons co-cultured with oligodendrocytes,112 primary CNS neurons co-cultured with oligodendrocytes and astrocytes,113 dendrite growth modelling,114 hippocampal axon compression injury,115 synapse formation in hippocampal neurons,116 embryonic neurons117 and P19-derived neurons co-cultured with mouse cortical neurons.118
These models allow for simultaneous segregation and connection between two or more cultures of neuronal and/or glial cells. However, this connection is equal in both directions, and some models require unidirectional connectivity to mimic specific neuronal circuitry. In these models discrete cellular connectivity in terms of inputs and outputs are used to infer not only elongation of neurites, but unidirectional control over axonal connectivity.119,120 Whilst grooves and channels orient process growth, it is a linear orientation with no directional selectivity, the neuronal processes grow from one chamber to the adjacent chamber and vice versa.114 In order to direct neural process growth in a single direction only, it is necessary to further optimise the design of the micro-channels between chambers. While Hattori et al.121 developed an asymmetrical scaffold to promote unidirectional connectivity by making the channel ascend as a slope in the undesired direction of growth, the selectivity and directional pressure was insufficient. An alternative design was later presented by Peyrinet al.122 which was similar to the basic microfluidic two-chamber device (Figure 1C), but featured asymmetrical micro-channels, aiming to create an oriented neural network. This tapered or ‘diode’ micro-channel design acted as a physical selector of directionality, with axons known to respond to physical cues in their microenvironment.
Two characteristics of axons in particular are exploitable for device design: axons can act as guide cues for other axons (with pioneer axons guiding follower axons through fasciculation and axonal bundling) meaning that larger channels accommodate more axons as soon as a pioneer axons finds the channel and enters; and axons react differently when meeting surfaces at different angles, either growing along the surface when aligned in parallel or deviating from their original direction when aligned in perpendicular, meaning sharp angles can be used to dissuade axonal growth whereas planar surfaces support axonal growth.123,124 To this end, Peyrinet al.122 designed channels that tapered in width from 15μm to 3μm in the desired direction of growth. This design imposed unidirectional axon connectivity with 97% selectivity.
Micro devices to support the culture of neuronal populations in order to mimic those circuits or connected populations found in vivo have now been well adopted into the neuroscience community. This has, to a large extent, been driven by multidisciplinary working, extending the capability of device design and manufacture whilst having the application focus of neural engineering. Despite the advances made, the majority of these in vitro models have, however, remained largely as 2D cell mono layers. In order to better mimic the in vivo tissue environment it is necessary to appreciate the 3D structure of the brain and how both neuronal and glial cells interact in 3D, leading to 3D in vitro neural cell models.
Neuronal and glial cell development in the CNS in vivo relies on complex cell-cell interactions in a 3D space.125 By focusing on 2D monolayer models, a vital component of in vivo brain structure and function is ignored. Hydrogels (such as collagen) are often used to present and maintain a 3D cell culture environment, with some systems further enabling delivery of therapeutics via the hydrogel matrix.126 By designing neural microdevices (as described above) that feature imbedded hydrogels to fill the cell culture area, researchers are extending the environment from the monolayer presented at the lower surface of the chambers into 3D.127–129 As well as the ability to interact in 3D space, the use of hydrogels also allows the extracellular environment to better mimic the stiffness of the brain, as the in vivo brain is soft, having a Young’s modulus of approx. 0.1-16kPa, compared to the 20-30GPa of tissue culture plastic or glass. Hydrogels are therefore considered to be much more accurate mimics of normal CNS tissue compared to the 2D growth surfaces of tissue culture plastic or glass,130 with neurons exhibiting faster network formation when grown on softer substrates,131,132
Cerebral organoids
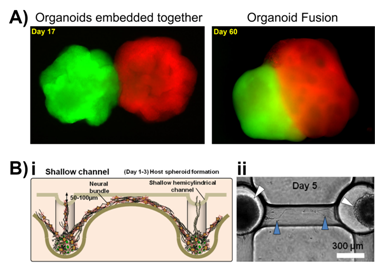
An example of the complexity that can be achieved by creating a 3D in vitro neural model comes in the form of organoid models, in this case cerebral organoids. These are supported 3D cell culture models which develop spatial regions with discrete identities that influence each other, similar to the early stages of the developing brain. Cerebral organoids can also exhibit cerebral regions that organise into various separated pyramidal identities as well as populations of outer radial ganglia,133 or can be fused together to generate a dorsal-ventral axis as shown in Figure 2A.134 These organoids represent the developing brain and as such have been used as in vitro neural models of neurodevelopmental disorders such as microcephaly, where the brain size is reduced. Lancaster et al.133 cultured neuroectoderm tissues in a spinning bioreactor to rapidly develop brain tissue and form a cerebral organoid. After 8-10days neural identity appeared, after 20-30days defined brain regions formed, and after two months tissues reached maximal size (size limit was hypothesised to be due to the lack of a circulatory system and issues with oxygen and nutrient exchange) forming complex heterogeneous tissues complete with forebrain, midbrain and hindbrain markers and boundaries, as well as tissues histologically similar to the cerebral cortex, choroid plexus, retina and meninges, that survived for up to 10months in the spinning bioreactor. Once the organoid was grown, RNA interference and patient-derived iPSCs were used to model microcephaly, which is usually difficult to recreate and model in mice or other in vitro models. Whilst these organoid models are morphologically and histologically similar to the developing brain, they cannot be (or at least have not been to date) functionally tested. The main criteria for analysis has focused on imaging techniques showing maturation of glial and neuronal cells, as well as their spatiotemporal organisation.133
Networked neurospheres
Choi et al.135 used this method to create an in vitro networked neurosphere model for Alzheimer’s disease. Neurospheres provide the means to present a semi-3D environment on a cluster of cells, with individual bodies sometimes referred to as ‘mini-brains’ when presenting differentiated neural populations. Choi et al. seeded concave micro wells with neural progenitor cells which self-aggregated to form uniform-sized neurospheres. These matured to connect to neighbouring neurospheres forming a multi-neural network by day 13. This model was shown to mimic the six organised horizontal layers of the cerebral cortex and was used to study the neurotoxicity of the protein amyloid beta (Aβ), known to play a part in causing Alzheimer’s disease. Adding Aβ tot eh network resulted in decreased cell viability and neurite degeneration.135
A similar neurospehere network produced by Jeong et al.136 to study signal transmission through the CNSas a result of the partial breakdown in this system seen in Alzheimer’s disease. Shallow (70µm) and deep (300µm) hemicylindrical channel networks between concave wells seeded with neural progenitor cells formed a self-aggregating network, Figure 2B. During this formation the cells differentiated into neuronal and glia cells that secreted laminin and formed an extracellular matrix (ECM) around the spheroids. Axonal signalling was recorded being transmitted between the spheroids, detected by Ca2+ flux imaging.136 Further models are summarised and reviewed in.137,138
An important consideration when designing an in vitro model of the brain is interstitial fluid flow. In vitro, the main roles of interstitial fluid are carried out by the cell culture medium, namely providing the cells with nutrients and removing waste during media changes. However, this culture medium in vitro is static, while in vivo the interstitial fluid flows throughout the brain, and this flow has numerous mechanical effects on the cells, as well as affecting communication between non-synaptic neurons.139 Reproducing the effects of this flow on cells in vitro will help models further mimic the in vivo environment, but interstitial fluid flow in the brain is very slow, measured at approximately 0.1-0.3 µL min-1,139 and reproducing/maintaining a flow of this speed can be a complicated process. Park et al. developed an osmosis-driven low-speed laminar flow technique to match this slow flow in vitro, allowing for testing of physiological flow on neuronal cells in vitro without exposing the cells to shear stress found with higher rates of flow.140 The inclusion of flow further increases the complexity of the in vitro model. The flow device was tested on a 2D culture of primary neural progenitor cells, which resulted in an increase in neurite length during differentiation when cultured with continuous flow compared to normal culture.141
By combining this osmotic pump with a networked neurosphere array, Park et al.142 created an in vitro model they termed a ‘brain-on-a-chip’ device. This model served not only as a mimic of the brain, but as a study of Alzheimer’s disease due to the addition of synthetic Aβ protein. Neurospheres were cultured both statically and in a dynamic model subjected to 0.15µL min-1 flow rate, with and without synthetic Aβ protein. Primary neural progenitor cells were seeded and cultured for 10 days to allow neurosphere formation, with toxic levels of Aβ protein added from day 7-10, allowing neurospheres to form in an environment more akin to Alzheimer’s disease. From days 4-10 the static neurospheres did not significantly change in size while the neurospheres in the dynamic flow environment increased in size. This suggests that flow may accelerate differentiation of neural progenitor cells (supported by higher levels of the neuronal marker β-III tubulin in the flow model), resulting in neurite outgrowth and synaptogenesis, increasing the neurosphere size. In addition, the treatment with Aβ had a much greater effect in the dynamic models, significantly reducing neurosphere viability and greater disruption of the neural networks compared to the static model.142 As with the previous 3D culture model employing neurospheres there was limited ability to test whether the neurons produced by this method were functional; only the differentiation status and morphology of the cells was analysed. This model represents a powerful platform for in vitro study of neurodegenerative disease, but without functionality testing via electrophysiology or other techniques, the resultant neurospheres network can only be so useful.
As fabrication and micro-manufacturing technology continues to improve, these permit more complex device designs to be realised in which to house and guide neural tissue engineering. The intricacy of these tissues is moving towards that of the central nervous system, albeit very slowly, with the enormity of the challenge highlighted by the plethora of cell types, their specific connectivity and regionality, and the 3D extracellular environment all playing pivotal roles. While in vivo models such as animal models have been a hallmark for attaining neural complexity in order to simulate a human brain and its accompanying disorders, these models may not necessarily the best option at present. Indeed, the prevalent nature of neurological disease is matched only by the persistent improvements in in vitro model technologies, moving from neuronal-only cultures, to neuronal/glial mixed cultures, to organised neural networks and circuits within microfluidic devices, to bio-artificial organs and organoids, modelling the CNS more accurately and efficiently with each leap in complexity.
Neurological disorders and diseases are debilitating conditions that currently have no cure. Difficulties of understanding the function an organ as complex as the brain, as well as the progression of disease and dysfunction contribute to our current stage of advancement in CNS research. These difficulties can be mostly abated by studying the brain in vitro at a pre-clinical level, but current pre-clinical assessment is insufficient to predict which treatments will work on human patients. One solution is to develop more efficient in vitro models presenting a high level of control and allowing the complexity to be increased to make the model more relevant. These models are low-cost and reproducible, combining cells with biomaterials and microfluidics to make lab-on-a-chip devices, which are the efficient in vitro models necessary for progress in research at a pre-clinical level, with the resulting data driving clinical trials in a more relevant direction, and contributing towards potential treatments for neurological or neurodegenerative diseases.
EPSRC-MRC Centre for Doctoral Training in Regenerative Medicine (EP/L015072/1).
The author declares no conflict of interest.

©2017 Köse-Dunn, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.