Advances in
eISSN: 2373-6402


Research Article Volume 9 Issue 2
1Department of Plant Breeding and Genetics, PMAS-Arid Agriculture University Rawalpindi, Pakistan
2Department of Plant Breeding and Genetics, University College of Agriculture and Environmental Sciences, Islamia University Bahawalpur, Pakistan
3Department of Arid Land Agriculture, King Abdul Aziz University, Saudi Arabia
4Department of Molecular and Cellular Biology, University of Guelph, Canada
Correspondence: Zahid Husain Shah, Department of Arid Land Agriculture, King Abdul Aziz University Jeddah, Saudi Arabia
Received: February 27, 2017 | Published: April 2, 2019
Citation: Khalil MU, Akram Z, Rana M, et al. Molecular marker assisted selection of promising wheat genotypes for high grain protein content. Adv Plants Agric Res. 2019;9(2):348-353. DOI: 10.15406/apar.2019.09.00447
Wheat contributes about 30% to the total cereals consumption across the globe. The high grain protein content of wheat ranks it as good quality. The genetic diversity has key importance in crop improvement programs. The purpose of present study was to evaluate 25 elite Pakistani (Rainfed) bread wheat varieties both at biochemical and molecular level so we can isolate the lines with broad genetic base and high quality controlling parameters. Maximum grain protein and ash contents were reported in genotypes NRL-0707, 9C037 and AUP-1059. Whereas, wet and dry gluten percentages were found highest in lines NRL-0707, AUR-0809 and DN-84 while lines NR-397 and NR-399 has shown the highest moisture content. However, both biochemical and molecular evaluation has segregated the lines NRL-0707, NR 397, 9C037, AUP-1052 and AUR-0809 as the most diverse lines for further utilization in wheat improvement programs.
Keywords: cluster analysis, genetic diversity, gluten, protein, SSR makers
Wheat is one of the most important cereal crop meeting one fifth caloric requirement of world population with per annum yield exceeding 680 million tonnes.1 Wheat has been a part of human diet since 8500 years, and currently it is providing 20% dietary protein.2 However, bread quality is a multipart aspect that relies on many factors. Most of the quality traits are usually complex and inherited quantitatively, especially milling yield, dough strength and extensibility. The quality and end-use suitability of wheat flour is associated with its grain protein composition.3 Environmental conditions strongly affect protein quantity while genotype and environment interaction determines its quality.4 Moreover, the grain storage proteins percentage can be referred as quality parameters as they determine the sustainability and plasticity of dough. However, the studies conducted during past few decades have provided a platform to understand the functionality of storage proteins and their links with allelic diversity.5 A cohesive and viscous proteinaceous compound, gluten, is prepared by storage proteins6 which means wheat proteins are analogous to other cereal proteins.6 When milled flour is mixed with water it attains the texture of dough, which retains air bubbles due to storage proteins. The structure of water insoluble gluten gets modify during various processing stages and it can be shaped into diversified kinds of food products.7 The selection efficacy of breeding germplasm has been enhanced by the development of functional markers linked to low and high molecular weight glutenin subunits.8,9
Moreover, the moisture percentage in the white flour and whole wheat flour has been reported as 11.3% and 11% respectively.10 On the other hand the moisture percentage has reported in fine flour and whole meal flour as 11.4% and 10.6% respectively11 which can be due to different methodologies of storage and processing. Moreover high moisture content is positively correlated with the fiber contents of thewheat grains,12 However, high moisture contents is important as it lowers the amount of dry solids as well as the shelf life of wheat product due to enhanced microbial activities.13 Similarly, high ash content indicates the high amount of minerals such as, calcium and iron which can be determined by combusting the sample of flour in the form of ashes.14 The ash content of wheat varies from 1.5 to about 2.0%. Breeding efforts for increased yield in wheat was considered by most breeders in the past while grain quality improvement remained neglected. In the present scenario, it is required to include some criteria in our existing breeding programs for the improvement of quality along with quantity.15 The efficiency of breeding and genetic conservation programs can be enhanced through exploitation of genetic diversity. It is now well understood that genetic diversity plays a vital role in achieving the ever increasing demand targets of wheat.16 Molecular analysis of plant genomes for the detection of polymorphism at DNA level with the utilization of various molecular techniques has revolutionized the assessment of genetic diversity.17 Efficiency of plant breeding programs can be enhanced greatly by the use of molecular marker technology that is an important area of biotechnology. The objective of current study was to make biochemical and molecular evaluations of elite Pakistani bread wheat varieties adapted to rainfed conditions. Moreover to segregate diverse genotypes from those that have been eroded genetically, so it can be decided about the improvement in their genetics via suitable breeding programmes.
In present study 25 selected genotypes from the “National Trials for Elite Entries (Rain-fed) 2014-15 (NTERF)” were evaluated for biochemical traits and genetic diversity.
Biochemical Evaluations
The germplasm was evaluated for biochemical contents like protein, moisture, ash and gluten. The obtained data were subjected to statistical analysis as advocated by Steel et al.18 The moisture percentage was calculated by simple air oven method, where 2 g flour sample was heated at 130˚C for one hour and the difference between the weights before and after heating was taken as moisture content. To determine ash content in wheat flour, known weight of flour was incinerated under controlled conditions and the left over residue was weighed. The percentage of ash was calculated based upon the original sample weight. However, protein content of the flour was determined by Dumus procedure while wet and dry gluten contents were determined by GLUTO-matic (Gultomatic-2200, Perten, Sweden) device.
Molecular evaluation
Seeds of these genotypes were grown in the growth chamber under strict controlled conditions. Tender leaf tissues from the young seedlings of all the genotypes were taken to extract DNA at the same time by using the method of Kang et al.19
PCR and Gel Electrophoresis
PCR was carried out in total 20 µl, contained 4 µl of total genomic DNA, 2 µl of each primer, 4 µl of dNTPs, 2 µl of Taq buffer with KCl, 1.2 µl MgCl2, 4.6 µl DEPC treated water and 0.2 µl of Taq DNA polymerase.20 The oligonucleotide sequences of the primers (forward and reverse) are given in Table 1. The amplification conditions for SSR analysis were as; denaturation for 1 minute at 93oC following 40 cycles, an annealing step at 60oC for 1 minute and an extension step at 72oC for 1 minute. The PCR amplified products using SSR were separated by gel electrophoresis and 2% high resolution agarose/TBE gel21 was utilized. Gels were stained with Ethidium Bromide and visualized under the UV light compartment and witnessed using the computer program UVI PhotoMW. For SSR analysis bands were scored as1 for present and 0 for absent. The 1-0 bivariate data matrix for each set of wheat lines based on the data of 2 SSR primer sets was used to construct dendrogram using computer program “Popgene32” version 1.31.22
Locus |
Right primer |
Left primer |
Xgwm4-4A |
TTCACCTCGATTGAGGTCCT |
GCTGATGCATATAATGCTGT |
Xgwm68-7B |
CTCCCTAGATGGGAGAAGGG |
AGGCCAGAATCTGGGAATG |
Xgwm99-1A |
GCCATATTTGATGACGCATA |
AAGATGGACGTATGCATCACA |
Xgwm102-2D |
TGTTGGTGGCTTGACTATTG |
TCTCCCATCCAACGCCTC |
Xgwm146-7B |
CTCTGGCATTGCTCCTTGG |
CCAAAAAAACTGCCTGCATG |
Xgwm164-1A |
TTGTAAACAAATCGCATGCG |
ACATTTCTCCCCCATCGTC |
Xgwm190-5D |
GTCCCACGTACCTTTG |
GTGCTTGCTGAGCTATGAGTC |
Xgwm257-2B |
CCAAGACGATGCTGAAGTCA |
AGAGTGCATGGTGGGACG |
Xgwm325-6D |
TTTTTACGCGTCAACGACG |
TTTCTTCTGTCGTTCTCTTCCC |
Xgwm332-7A |
AGTGCTGGAAAGAGTAGTGAAGC |
AGCCAGCAAGTCACCAAAAC |
Table 1 List of SSR primers used for molecular evaluation
Biochemical evaluation
Protein content
Wheat flour with protein content 11 % or more has better bread making quality. The analysis of variance for protein contents among these genotypes has shown significant difference (Table 2). Mean values for protein contents varied between 12.12 (genotype NRL-0707) to 14.24 % (PR-105) as shown in Table 3.
Sr. No. |
Traits |
DF |
Genotypic mean squares |
Error mean squares |
1 |
Protein content |
24 |
0.59728** |
0.02475 |
2 |
Moisture content |
24 |
1.04960** |
0.07385 |
3 |
Ash content |
24 |
0.00773** |
0.00016 |
4 |
Gluten content wet |
24 |
4.89185** |
0.09903 |
5 |
Gluten content dry |
24 |
0.43672** |
0.00128 |
Table 2 Analysis of variance for biochemical characters
** Highly significant
Sr. No. |
Genotypes |
Protein (%) |
Moisture (%) |
Ash (%) |
Wet gluten (%) |
Dry gluten (%) |
1 |
PR-104 |
12.45 |
9.74 |
1.36 |
21.2 |
8.06 |
2 |
PR-105 |
12.12 |
9.92 |
1.29 |
20.41 |
8.05 |
3 |
AUR-0809 |
12.4 |
9.63 |
1.32 |
22.66 |
8.13 |
4 |
AZRC-2 |
12.32 |
10.34 |
1.34 |
19.52 |
7.76 |
5 |
NRL-0707 |
14.24 |
10.23 |
1.42 |
24.74 |
9.16 |
6 |
DN-84 |
12.38 |
11.31 |
1.37 |
21.46 |
8.26 |
7 |
VO8BT022 |
12.31 |
9.96 |
1.28 |
20.2 |
7.97 |
8 |
V-11183 |
12.3 |
10.45 |
1.31 |
21.5 |
8.19 |
9 |
V-08314 |
12.19 |
9.85 |
1.32 |
19.55 |
7.79 |
10 |
NR-399 |
12.41 |
11.43 |
1.21 |
20.44 |
7.93 |
11 |
NR-400 |
12.22 |
11.06 |
1.28 |
19.66 |
7.87 |
12 |
9C037 |
13.22 |
10.39 |
1.38 |
19.08 |
7.24 |
13 |
DH-31 |
12.75 |
9.65 |
1.26 |
18.73 |
7.05 |
14 |
10C033 |
12.18 |
10.76 |
1.3 |
20.64 |
8.08 |
15 |
06 FJS3013 |
12.64 |
11.35 |
1.33 |
20.4 |
8.06 |
16 |
NR-419 |
12.52 |
9.79 |
1.28 |
19.67 |
7.75 |
17 |
NR-420 |
12.37 |
10.56 |
1.29 |
20.33 |
8.13 |
18 |
NIA-MB-2 |
12.25 |
10.46 |
1.31 |
21.48 |
8.22 |
19 |
NR-403 |
12.68 |
9.7 |
1.28 |
19.77 |
7.65 |
20 |
AUP-1052 |
12.37 |
9.96 |
1.27 |
19.59 |
7.89 |
21 |
AUP-1059 |
13.08 |
10.25 |
1.43 |
20.64 |
7.85 |
22 |
Pirsabak-0 |
12.56 |
9.84 |
1.34 |
19.56 |
7.78 |
23 |
NARC-2009 |
12.33 |
10.49 |
1.3 |
20.1 |
8.04 |
24 |
Chakwal-50 |
12.41 |
10.34 |
1.26 |
19.67 |
7.93 |
25 |
NR 397 |
12.19 |
11.54 |
1.27 |
19.2 |
7.59 |
Table 3 Mean estimated values of various biochemical analyses
Moisture content
The analysis of variance for moisture content has revealed significant difference among all genotypes (Table 2). Mean values for moisture content ranged between 9.63 (AUR-0809) to 11.54% (NR-397) as represented in Table 3.
Ash content
Significant differences were witnessed among all wheat lines for ash percentage (Table 2) with mean values ranging between 1.21 (NR-399) to1.43% (AUP-1059) as mentioned in Table 3.
Wet gluten content
Gluten in wheat is considered to be a rough estimate of its total protein content. The gluten content of flour increased with the increase in its protein content. The gluten contents estimated among all these genotypes were significantly different. A wide range of mean values was seen for wet gluten varying between 18.73 (DH-31) to 24.74% (NRL-0707) as mentioned in Table 3.
Dry gluten content
The differences in gluten content may be ascribed to the differences in parents of the selected wheat lines, variation in climatic conditions and differences in cultural practices. Analysis of variance for dry gluten content in Table 2 has represented highly significant differences among all genotypes. The mean values were observed in a range between 7.06 % (DH-31) to 9.16% (NRL-0707) as indicated in Table 3.
Molecular evaluation
For the detection of DNA polymorphism among selected genotypes a total set of 10 primers was used as shown in Table 1. Collectively, these primers had amplified 532 bands in all these genotypes with size ranging from 50bp to 1000bp. Maximum number of bands, 111 were traced by primer Xgwm332-7A, while the lowest, 25 by the primer Xgwm102-2D. Different primers revealed variation in their ability to detect polymorphism, primers Xgwm4-4A (Figure 1d), Xgwm68-7B, Xgwm102-2D, Xgwm146-7B, Xgwm164-1A, Xgwm190-5D (Figure 1c) and Xgwm332-7A (Figure 1b) amplified all genotypes. While primers, Xgwm257-2B and Xgwm325-6D (Figure 1a) have shown amplification in 23 genotypes. However, only Xgwm99-1A has amplified 24 genotypes.
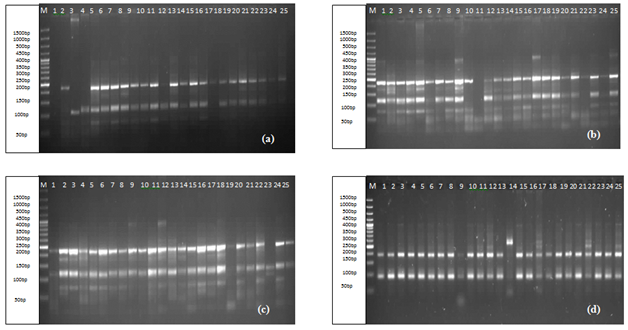
Figure 1 Amplification patterns of SSR primers, Xgwm325-6D (a) Xgwm332-7A; (b) Xgwm190-5D; (c) Xgwm4-4A; (d) analyzed using the computer program UVI PhotoMW.
Dendrogram interpretation
On the basis of genetic distances data dendrogram was obtained which has grouped these genotypes in seven clads as indicated in Figure 2. Three genotypes PR-104 (V1), NRL-0707 (V5) and NR-397 (V25) are the part of same clad. The overall genetic distance of this cluster lies between 2.00 to 2.65%. Among these genotypes, line PR-104 and NRL-0707 has shown least genetic distance of 2.00% and are the part of same sub group. However, the line NR-397 was the most diverse line in this cluster with a genetic distance of 2.65%. Moreover this clad is totally isolated as compared to the other clusters in the dendrogram. Similarly, 9C037 (V 12) and AUP-1059 (V 21) were also genetically distinct as compare to other genotypes as they were separated from other clads of dendrogram.
Ahmad et al.23 Khan et al.24 Rharrabti et al.25 Maghirang et al.26 and Ikhtiar and Alam15 reported the presence of protein content from 12- 15% in their studies which are in complete agreement with our findings. Moreover, flour with high protein and cakes with low protein are more expensive usually. Hence, there is a good correlation between bakery performance of flour and protein content. Pearson et al.26 Ahmad et al.23 Samman et al.27 and Ikhtiar & Alam15 noted moisture content of different wheat genotypes from 8 to 12%. We also reported analogous results in our study. It is miller’s interest to keep moisture contents in wheat below 14%, as above this moisture content flour becomes unstable due to exponential microbial growth which produces bad odor in flour and deteriorate its quality. Moreover, the ash amount in the wheat kernel is 1 to 2%, with 0.35% concentrated in endospermic region that constitute 80% part of kernel.15,23,25 This was in complete agreement with our results. Ash content is a measure of the amount of impurity present in wheat flour. Hence, it is the objective of the millers to separate non-endospermic part to get high starch flour with low ash contents.
Analogous to Oak et al.28 and Khan et al.29 we also reported wet gluten from 18 to 25% that is quantitative measure of the protein responsible for gluten formation which not only enhances dough mixing but also improves baking quality. Furthermore, the mean values for dry gluten content were observed from 7.06 to 9.1%. which is according to the findings of Ahmad et al.23 Anjum et al.30 Oak et al.28 and Samman et al.27 However, Khan et al.29 found gluten content from 10.49 to 13.60%. On the other hand, Bonomi et al.7 found that there is no co-relation among the concentration of gluten, and ash. But our findings revealed a positive co-relation between moisture and gluten. Biochemical study suggested that lines NRL-0707, 9C037 and AUP-1052 were found rich in grain protein and ash content. Gluten content were found highest in lines NRL-0707, AUR-0809 and DN-84 while lines NR 397 and NR-399 performed best for moisture content in wheat (Table 3). Moreover, the genotypes those were rich in proteins contents also shown high gluten contents as indicated in Table 3. However, cluster analysis recommended lines NRL-0707, NR 397, PR-104, 9C037 and AUP-1052 as most diverse lines among all. This indicated that these lines would result well in further breeding programs due to their maximum genetic diversity. The fall outs attained in this molecular study are in settlement with the genetic diversity estimation of wheat conducted by Borner et al.31 Zhang et al.32 In a previous study conducted by Bryan et al.33 Huang et al.34 Ni et al.35 and Nisar et al.36 who utilized SSR markers for genetic diversity estimation of grain protein in wheat. From both biochemical and molecular evaluations we can conclude that the lines NRL-0707, NR 397, 9C037, AUP-1052 and AUR-0809 (Figure 3) are lines that are superior in their quality and genetically less eroded, hence they can be used further in breeding programmes.37‒47
Along with quantity emphasis should also be given on the quality parameters of wheat that are concerned directly with the health of human being. So, breeders should also target the nutritional aspects of wheat when exploiting the diversity for broadening the genetic base of their germplasm. Hence our conducted study will help the breeders to formulate further hypothesis by linking nutritional parameters with genetic diversity of wheat.
None.
The authors declare that there is no conflict of interest.

©2019 Khalil, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.