Advances in
eISSN: 2373-6402


Research Article Volume 9 Issue 1
Plant Genetic Resources Department of Desert Research Center, Matariya, Cairo, Egypt
Correspondence: RMA Khalil, Plant Genetic Resources Department, Desert Research Center, Matariya, Cairo, Egypt,
Received: December 26, 2018 | Published: January 17, 2019
Citation: Khalil RMA, El zayat MAS. Molecular characterization of some Brassica species. Adv Plants Agric Res. 2019;1(3):112-119. DOI: 10.15406/apar.2019.09.00420
Nineteen ISSR were used to investigate polymorphism among five Brassica species. ISSR revealed326 total bands the highest (89% polymorphism). 30SRAP primer pairs revealed 480bands with polymorphism (93%). ISSR and SRAP, both were effective in studying genetic diversity in Brassica. Based on similarity matrix of overall analysis (ISSR and SRAP), the dendrogram was constructed and separated the five Brassica into two main clusters. Results showed that the length of the MYB28 gene for glucosinolates production at the fragment of 1257bp.This information can be used in the future for breeding and improvement programs and helps in national conservation programs.
Keywords: Brassica, ISSR, SRAP, glucosinolates and MYB28gene
Brassica genus (Brassicaceae) contains proteins, vitamins, minerals, glucosinolates and phenols. Glucosinolates contribute to reducing the risk of cancer.1 That activity is mainly related to the presence of isothiocyanates and nitralates. Most species are of great economic importance since most of them are edible, a source of high-value edible oil, fodder, Which helps to protect the food supply by expanding the range of genes available in the face of the challenges of the lack of agricultural production, and is of great medical importance such as: white mustard, black, brown, yellow, cabbage and broccoli help in the treatment of chronic, cough, asthma, chest pains, heart disease, headache, compresses the pain of rheumatism, joints, diuresis, stimulates the metabolism, clears the appetite, treats tumors, regulates blood sugar and prevents cerebral palsy. Molecular markers are effective tools for revealing genetic diversity, inter-sequence simple repeat (ISSR), is useful for detecting genetic polymorphisms among genotypes by generating a large number of markers that target multiple microsatellite loci distributed across the genome.2 ISSR is a technique that overcomes most of other techniques limitations. This technology has been used to DNA fingerprint a wide range of species of medicinal plants, phylogeny and the geographic origin of some plant species as Brassica napus.3 Sequence-related amplified polymorphism (SRAP) is a new and useful molecular marker system for tagging and mapping in B. oleracea.4 The SRAP system is easy to perform, gives stable and moderate yields and a high proportion of co-dominance, and ultimately simplifies the cloning and sequencing of target segments. The SRAP technique is an effective tool for constructing genetic and linkage maps,5 carrying out comparative genomics and analyzing genetic diversity because sequences are amplified against open reading frames, and sequences near the centromeres and telomeres of genomes are only rarely amplified. The SRAP technique has been applied successfully in studies of sweet potato, rape, and Chinese cabbage and brocoli.6-8 Although there are many genetic studies related to plant traits, there are few in terms of identifying genes and their characterization, especially specific of Brassica for the main active substance such as glucosinolates .There is a great challenge to be addressed in the inventory of genetic variation that helps us to improve crops. Glucosinolates, in turn, are resistant to pests and microbes, as well as the industrial significance of brassica in the work of bombs in wars, thus preserving genetic diversity in gene banks and characterizing them and defining the important genes that we can transfer to important crops to be used as food and source of important effective substances. (MYB28) gene was found to participate in regulating the aliphatic GSL biosynthesis glucosinolates and have been successfully identified in several plant species including Arabidopsis thaliana,9 Brassicajuncea,10 and B. rapa.11 The objectives of this study were to investigate the level of genetic diversity and relationships in Brassica by molecular markers and provide valuable information for detecting glucosinolates gene.
Five Brassica species were collected from the Gene Bank of North Sinai Station, Desert Research, Egypt during February 2015-2017, as shown in (Figure 1) (Table 1).

Figure 1 The five Brassica species collected from the Gene Bank of North Sinai Station, Desert Research, and Egypt.
No |
Scientific name |
1 |
Brassica tournefortii |
2 |
Brassica nigra L. |
3 |
Brassica rapa L. |
4 |
Brassica Juncea L. |
5 |
Brassica carentia |
Table 1 Taxa of the five studied Brassica species
DNA extraction
A total of 25 young fresh leaves from each of the 20 annual ryegrass plants were randomly collected and extracted using the DN easy Plant Mini kit (QiagenInc, Valencia, CA, USA). The quality and concentration of the DNA were confirmed by electrophoresis on 0.8% agarose gels with the standardized lambda DNA size markers.
ISSR analysis
Finally, 15 primers (Life Technologies) (Table 2) were selected for ISSR analysis. The PCR amplification comprised a total volume of 20mL containing 3mL 20ng/mL DNA, 0.2mL Taq DNA polymerase, 1.4mL 2.5 mMdNTPs, 3mL 10mM primer, 1.4mL 25mM Mg , 2.5mL 10X buffer, and 8.3mL ddHO. PCR amplification was carried out on the Eppendorf PCR instrument and the reaction program comprised an initial 5 min at 94°C; 35 cycles of 45 s at 94°C, 45 s annealing at 50°C and a 90-s extension at 72°C; ending with a final extension of 5 min at 72°C and storage at 4°C. Amplified products were electrophoreses on 6% denaturing poly acrylamide gel, which was silver stained and photographed gels and visualized with the Gel Documentation System(Bio-Rad® Gel Doc-2001) (Germany).
Primer name |
Sequence |
Primer name |
Sequence |
HB01 |
(CAA) 5 |
844B |
(CT) 8 GC |
HB02 |
(CAG) 5 |
HB9 |
(GT) 6 GG |
HB04 |
(GACA) 4 |
HB10 |
(GA) 6 CC |
17898A |
(CA) 6 AC |
HB11 |
(GT) 6 CC |
17898B |
(CA) 6 GG |
HB12 |
(CAC) 3 GC |
17899A |
(CA) 6 AG |
HB14 |
(CTC) 3 GC |
17899B |
(CA) 6 GG |
HB15 |
(GTG) 3 GC |
844A |
(CT) 8 AC |
814 |
(CT) 8TG |
807 |
(AGA) 4GT |
HB13 |
GAC)3 GC |
HB8 |
(GA)6 GG |
|
|
Table 2 ISSR primer sequences
SRAP fingerprinting
Thirty selected pairs of primers (Life Technologies) (Table3) from 60 different primer combinations were used for detecting polymorphism in open reading frames (ORFs). Each 20-μLPCR mixture consisted of 1. 0U Taq DNA polymerase, 6X PCR buffer, 0.6m MdNTP, 0.35μM primer, 1.5 mM Mg, and 25-200ng template DNA. Thermal cycling (Biometra T1 Thermo cycle) started with 5min at 94°C for initial denaturing, and 5 cycles of 30s at 94°C, 30s at 35°C, and 45 s at 72°C, followed by 40 cycles of 30s at 94°C, 30 s at 4°C and 45 s at 72°C. The last cycle was followed by a 7-min extension at 72°C. Amplified products were analyzed on 2% (w/v) gels and visualized with the Gel Documentation System (Bio-Rad® Gel Doc-2001) (Germany) (Table 3).
Primer name |
Forward primers |
Primer name |
Reverse primers |
Me1 |
TGAGTCCAAACCGGATA |
Em1 |
GACTGCGTACGAATTAAT |
Me2 |
TGAGTCCAAACCGGAGC |
Em2 |
GACTGCGTACGAATTTGC |
Me3 |
TGAGTCCAAACCGGAAT |
Em3 |
GACTGCGTACGAATTGAC |
Me4 |
TGAGTCCAAACCGGACC |
Em4 |
GACTGCGTACGAATTTGA |
Me5 |
TGAGTCCAAACCGGAAG |
Em5 |
GACTGCGTACGAATTAAC |
Me6 |
TGAGTCCAAACCGGACA |
Em6 |
GACTGCGTACGAATTGCA |
Me7 |
TGAGTCCAAACCGGACG |
Em7 |
GACTGCGTACGAATTCAA |
Me8 |
TGAGTCCAAACCGGACT |
Em8 |
GACTGCGTACGAATTCTG |
Me9 |
TGAGTCCAAACCGGAGG |
Em9 |
GACTGCGTACGAATTCAG |
Me10 |
TGAGTCCAAACCGGAAA |
Em10 |
GACTGCGTACGAATTCAT |
Me11 |
TGAGTCCAAACCGGAAC |
Em11 |
GACTGCGTACGAATTCTA |
Me12 |
TGAGTCCAAACCGGAGA |
Em12 |
GACTGCGTACGAATTCTC |
DN06 |
TGAGTCCAAACCGGTAA |
Em13 |
GACTGCGTACGAATTCTT |
DN07 |
TGAGTCCAAACCGGTCC |
Em14 |
GACTGCGTACGAATTGAT |
DN08 |
TGAGTCCAAACCGGTGC |
Em15 |
GACTGCGTACGAATTGTC |
DN09 |
TGAGTCCAAACCGGTCA |
Em16 |
GACTGCGTACGAATTCGA |
DN10 |
TGAGTCCAAACCGGGCT |
Em 17 |
GACTGCGTACGAATTAGC |
DN11 |
TGAGTCCAAACCGGTAG |
Em 18 |
GACTGCGTACGAATTGAG |
DN12 |
TGAGTCCAAACCGGTGT |
Em 19 |
GACTGCGTACGAATTGCC |
Table 3 The forward and reverse SRAP primer in this study
Statistical analysis
Similarity matrix was developed by the statistical package for social science program SPSS based on combined analysis of overall molecular and biochemical markers.
Detection of gene polymerase chain reaction (PCR) and sequencing
PCR was conducted using forward and reverse primers, which were designed based on these quence of MYb28 obtained from the Gen Bank (BLAST). These quinces of the primers were 5'-GGGACCATCACACAATTCATTTCTC-3' (forward) and 5'-' (reverse). A mixture of 20μL solution consisting of 10xEx Taq polymerase buffer, 2m MMgCl2, 200u Md NTPs, 25p mol primers, 1UTaqpolymerase and distilled water was used for each PCR reaction. The PCR program was seton 94°C for 2minutes pre heating continued with 35cycles consisting of 1minute denaturation at 94°C, 1minutes annealing at 56oC, and90 seconds extension at 72°C. The PCR product was visualizedon1,5% agarose gel and subjected to 100 volts for 1hr and then photographed using UV gel documentation system, UVP corporation- UK.
Purification of PCR product and sequencing: PCR products were purified using High Pure PCR Product purification Kit (Roche) and sequenced (MWG, Germany).
ISSR analysis
Nineteen preselected ISSR primers were used in the present investigation to study the genetic relationships among the five studied Brassica species as shown in ( Figure 2) and (Table 4) .326 total bands, 36 monomorphic and 290 polymorphic distinct bands (89% polymorphism) were generated by the nineteen ISSR primers. The results showed that 17898B, 17899A, HB1, HB4, HB9, HB11 and HB15 primers were highly polymorphic (100% polymorphism), while HB13 generated the lowest polymorphism (74%). These results agreed with the results obtained by Khalil)2010( who used fifteen preselected ISSR primers to identify eleven species of Brassicacea and produced72 polymorphic distinct fragments (56.2% of polymorphism). Those results gave another dimension to detect the genetic variability in accurate differentiations and demonstrated significant levels of variation. Moreover, Santos et al., (2016) studied ISSR primers and were capable of detecting genetic polymorphism among Mimosa caesalpiniaefolia. A total of 7 ISSR primers generated 52.7% polymorphism.
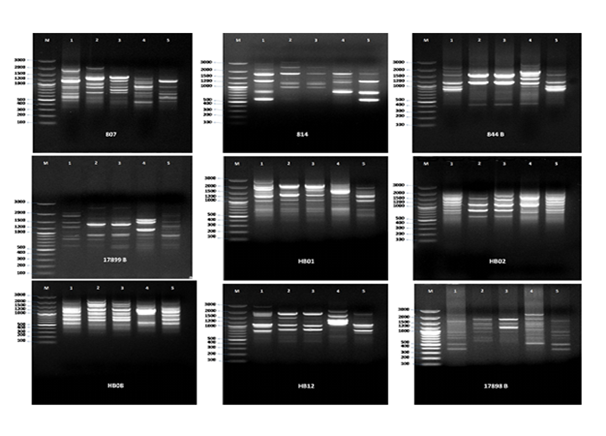
Figure 2 ISSR primers of five Brassica species*. ** (M) = Maker, *(Brassica tournefortii (Gouam)) = 1, (Brassica nigra L.)=2, (Brassica rapa L.) = 3, (Brassica Juncea L.) = 4, (Brassica carentia) = 5.
S. No |
Primer code |
Monom orphic bands |
Polymorphic bands |
Unique bands |
Total amplified bands |
Polymorphism percentages |
1 |
807 |
2 |
18 |
7 |
20 |
90% |
2 |
814 |
1 |
15 |
7 |
16 |
94% |
3 |
844A |
3 |
12 |
4 |
15 |
80% |
4 |
844B |
1 |
14 |
5 |
15 |
93% |
5 |
17898A |
1 |
14 |
5 |
15 |
93% |
6 |
17898B |
0 |
17 |
3 |
17 |
100% |
7 |
17899A |
0 |
13 |
3 |
13 |
100% |
8 |
17899B |
2 |
15 |
6 |
17 |
88% |
9 |
HB1 |
0 |
13 |
3 |
13 |
100% |
10 |
HB2 |
5 |
9 |
0 |
14 |
64% |
11 |
HB4 |
0 |
20 |
7 |
20 |
100% |
12 |
HB8 |
1 |
14 |
2 |
16 |
88% |
13 |
HB9 |
0 |
8 |
1 |
8 |
100% |
14 |
HB10 |
2 |
24 |
9 |
26 |
92% |
15 |
HB11 |
0 |
14 |
1 |
14 |
100% |
16 |
HB12 |
2 |
15 |
4 |
17 |
88% |
17 |
HB13 |
6 |
17 |
3 |
23 |
74% |
18 |
HB14 |
1 |
13 |
6 |
14 |
93% |
19 |
HB15 |
0 |
21 |
7 |
21 |
100% |
|
Total |
27 |
286 |
83 |
329 |
87% |
Table 4 Primer codes, length range (bp), monomorphic bands, polymorphic bands, total amplified bands and polymorphism percentages of nineteen ISSR primers amongfive Brassicas pecies
SRAP analysis
Thirty pairs of preselected SRAP primers were used in the present investigation to study the genetic relationships among the five studied species of Brassica as shown in (Figure 3) and (Table 5) (Table 6). 480 total bands, 32 monomorphic bands and 448polymorphic distinct bands (93% polymorphism) were revealed by the SRAP primers. The results showed that EM1RXme8F, EM6RXme5F, Em6RXme11F, EM10RXDN6F, EM10RXDN7F, Em10RXDN12F, Em13RXDN12F, Em17RXDN12F and Em18RXDN11F primer pairs were highly polymorphic (100% polymorphism). On the other hand, EM6RXDm8Fprimer pair revealed the lowest polymorphism (75%). Polymorphic information content (PIC) and Marker indexes (MI) were equal between ISSR and SRAP markers (Table 6). These results agreed with the results obtained by Ahmad et al., (2014) who studied the genetic diversity and relationships among B. napus accessions using SRAP markers, which preferentially amplify open reading frames. Using 20 SRAP primers, a total of 60 spring-type B. napus accessions revealed 162 polymorphic fragments. Cluster analysis displayed five major groups. The clustering pattern mostly supported their respective pedigree and characteristic traits.
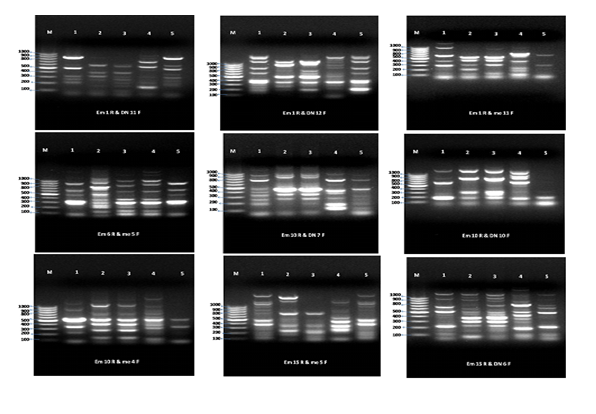
Figure 3 SRAP primers amongfive Brassica species*.**(M) = Maker, *(Brassica tournefortii (Gouam)) = 1, (Brassica nigra L.) =2, (Brassica rapa L.) = 3, (Brassica Juncea L.) = 4, (Brassica carentia) = 5.
NO |
Primer code |
Monom orphic |
Total polymorphic |
Unique |
Total amplified |
Polymorphism |
1 |
Em1RXDN11F |
1 |
12 |
3 |
16 |
94% |
2 |
Em1RXDN12F |
1 |
10 |
9 |
20 |
95% |
3 |
EM1RXme8F |
0 |
7 |
6 |
13 |
100% |
4 |
EM1RXme11F |
1 |
12 |
7 |
13 |
93% |
5 |
EM2RXme12F |
1 |
10 |
7 |
11 |
90% |
6 |
EM6RXDm8F |
3 |
9 |
3 |
12 |
75% |
7 |
EM6RXme5F |
2 |
17 |
7 |
19 |
100% |
8 |
Em6RXme11F |
0 |
18 |
4 |
18 |
100% |
9 |
EM8RXme1F |
1 |
8 |
1 |
9 |
88% |
10 |
EM9RXme1F |
1 |
16 |
5 |
17 |
94% |
11 |
EM10RXDN6F |
0 |
17 |
3 |
17 |
100% |
12 |
EM10RXDN7F |
0 |
20 |
6 |
20 |
100% |
13 |
EM10RXDN8F |
3 |
10 |
6 |
13 |
77% |
14 |
Em10RXDN10F |
2 |
10 |
4 |
12 |
83% |
15 |
Em10RXDN12F |
0 |
17 |
4 |
17 |
100% |
16 |
Em10RXme4F |
1 |
16 |
5 |
17 |
94% |
17 |
Em12RXDN10F |
1 |
15 |
8 |
16 |
94% |
18 |
Em12RXDN12F |
1 |
17 |
3 |
18 |
94% |
19 |
Em13RXDN12F |
0 |
14 |
4 |
14 |
100% |
20 |
Em15RXDN6F |
2 |
19 |
5 |
21 |
90% |
21 |
Em15RXme10F |
3 |
24 |
0 |
27 |
89% |
22 |
Em15RXDN7F |
1 |
18 |
3 |
19 |
95% |
23 |
Em15RXDN9F |
1 |
8 |
9 |
9 |
88% |
24 |
Em15RXme11F |
1 |
8 |
1 |
9 |
88% |
25 |
Em17RXDN10F |
0 |
18 |
6 |
18 |
100% |
26 |
Em17RXDN12F |
0 |
12 |
3 |
12 |
100% |
27 |
Em18RXDN11F |
0 |
10 |
5 |
10 |
100% |
28 |
Em18RXDN12F |
2 |
10 |
1 |
12 |
83% |
29 |
Em20RXDN10F |
1 |
16 |
1 |
17 |
94% |
|
Total |
30 |
398 |
129 |
485 |
82% |
Table 5 Primer codes, length range (bp), monomorphic bands, polymorphic bands, total
Parameters for marker efficiency |
ISSR |
SRAP |
Number of individuals |
19 |
30 |
Total number of bands (L) |
326 |
480 |
Polymorphic bands (p) |
290 |
448 |
Number of loci/assay unit (nu) |
17.1 |
16 |
Total number of effective alleles (Ne) |
1213 |
1975.8 |
Average number of polymorphic bands/assay unit(np/U) |
15.26 |
14.93 |
Polymorphic information content (PIC) |
0.97 |
0.97 |
Fraction of polymorphic loci (β) |
0.88 |
0.93 |
Assay efficiency index (Ai) |
63.8 |
65.8 |
Effective multiples ratio (E) |
15.26 |
14.93 |
Marker index (MI) = Hav × MR |
14.82 |
14.62 |
Table 6 Relative efficiency of molecular markers in determining polymorphism among five
Additionally, Yu12 used SRAP and ISSR markers to assess the genetic diversity within and among 15 natural populations of Stip abungeana from the Loess Plateau of China. Using 15 SRAP primers, 504 (99.80%) polymorphic loci were detected, and 372 polymorphic loci (96.12%) were identified using 15 ISSR primers. The results showed that genetic variation within populations is lower than that among populations. The results demonstrated that both SRAP and ISSR markers are effective and reliable for assessing the genetic diversity of S. bungeana. In addition, these data inform conservation and breeding strategies for S. bungeana. Besides, Ma et al.,13 detected the genetic relationships of different Cotoneaster schantungensis natural populations and genetic diversity analysis based on SRAP markers. Twelve pairs primers were selected out, 93 bands were amplified, of which 91 bands (97.85%) were polymorphic. Guenni et al., (2016) used SRAP markers preferentially to amplify open reading frames and to study the genetic diversity of 43Tunisian pistachio accessions. Using seven SRAP primer pairs, a total of 78 markers revealed (95.12%) that will help to design future conservation and breeding strategies concerning this species.
Genetic and phylogenetic relationships
The generated ISSR and SRAP profiles were further used to assess similarities among the studied cultivars. The estimation of genetic distance was accomplished using a computer software (NTSYS-PC version 1.8) based on the formula originally proposed by Nei and Li (1979). The results are presented in (Table 7). Genetic distances ranged from (.385 to .762) in ISSR and SRAP Marker. The lowest genetic distance was found between Brassica tournefortii and B. rapa. The highest genetic distances were registered between B. nigra and B. rapa. The genetic distances were supported by the resulting dendrogram (Figure 4). Dendrogram revealed two main clusters, the first cluster splatted into two sub-clusteres included B. tournefortii and B. rapa. The second cluster included B. nigra and B. rapa in one sub cluster and B. junceain another The use of ISSR and SRAP marker is strongly supported by many studies dealing with the usefulness of such markers for investigating Brassica diversity. The current research is one of the attempts to use molecular markers in investigating the genetic relationships among various local genotypes geranium grown in desert Egypt.
|
Matrix File Input |
|||
Case |
Brassica tournefortii |
Brassica nigra |
Brassica rapa |
Brassica juncea |
Brassica tournefortii |
||||
Brassica nigra |
0.386 |
|||
Brassica rapa |
0.385 |
0.762 |
||
Brassica juncea |
0.462 |
0.457 |
0.48 |
|
Brassica carentia |
0.675 |
0.393 |
0.4 |
0.445 |
Table 7 Comparison by similarity of relationships Brassica spp. genotypes as revealed by ISSR, SRAP marker systems
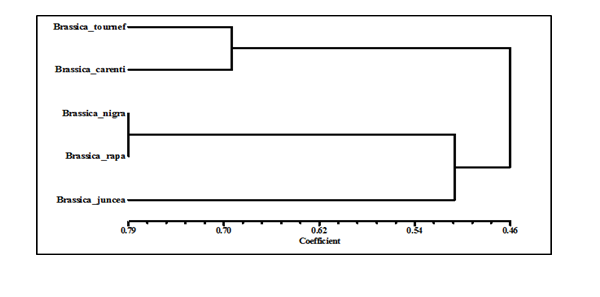
Figure 4 An un weighted pair-group method with arithmetic averages (UPGMA) dendrogram of genetic relationships among five genotypes of Brassica obtained using ISSR and SRAP combined data.
MYB28 gene detection
The PCR product using specific primer of MYB28 gene indicated that the appearance of one band for each with fragment size 1257bp as shown in (Figure 4) (Figure 5). These results agreed with the results obtain Ling et al.,14 who isolated and characterized MYB28gene from B. oleraceae.
Analysis of the MYB28 gene nucleotide sequence alignment
Sequencing and BLAST analysis showed that the length of TDC fragment shares high homology with the other known MYB28 as shown in (Figure 6) (Figure 7). Homology search results in Gene Bank (NCBI) showed that MYB28 nucleotide had high identity to other plants such as B. oleraceae (94% identities, accession number (AB702694.1) and Raphanussativus (89% identities, accession number XM018625413.1).The highest identities was 99% for B. rapa.
Phylogenetic analysis of DNA sequence of TDC gene
Phylogenetic analysis was done by aligning DNA sequences using software Clustal X software to construct a phylogenetictree (Figure 8). MYB28 gene among Brassica five species. Brassica tournefortii, B. oleraceae B. juncea , B. rapa and Raphanus sativus were grouped together and B. nigra and B. crentia were closer and group in another cluster.
ISSR and SRAP used effectively to estimate genetic diversity among investigated five species of Brassica and may help further inbreeding programmers. Glucosinolates gene explorations are required to have better understanding of the presence of genetic variability in Brassica for improvement of other relative important crops.
The authors declared there is no conflict of interest.

©2019 Khalil, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.