Advances in
eISSN: 2373-6402


Research Article Volume 9 Issue 1
Department of Biological Science, Karakoram International University, Pakistan
Correspondence: Tika Khan, Department of Biological Science, Karakoram International University, Gilgit, Pakistan
Received: July 16, 2018 | Published: January 16, 2019
Citation: Gul Z, Khan IA, Kiran C, et al. Comparative genomic, proteomic and morphological studies in two indigenous artemisia species collected from high mountains of Karakorum and himalaya in Gilgit Baltistan. Adv Plants Agric Res. 2019;9(1):106-110. DOI: 10.15406/apar.2019.09.00419
Artemisia is a wild widely distributed medicinally important genus. Thirty-eight species have been reported from Pakistan. Genetic diversity between A. gemilini and A. bravifolia has been estimated using genetic distances (GD) out of genomic, proteomic and morphological documentation. Samples were collected from 6 locations ranging between 1400 and 5500 m above mean sea level. Both species showed significant differences for morphological characters. Total crude protein concentration in leaves of Artemisia gemilini (19.4 %+0.46, range 18.5-20.7 %) and Artemisia bravifolia (19.3 %+0.53, range 17.6-20.7 %). SDS-PAGE revealed specific protein bands only in A. gemilini. Total genomic DNA extracted from bark using optimized isolation protocol. Twenty-eight RAPD primers amplified 125 fragments resulted 4.5 fragments per primer. Eight primers exhibited 100 % genetic distance. Genomic and proteomic bivariate (1-0) data matrix analyzed using UPGMA for SDS-PAGE and RAPD amplification ranged from 0-75% and 1-100% GD respectively. Extraction protocol and results from the study can be useful for future reference, taxonomic revision, conservation, hybridization programs and improvement in cut tree optimization of dendrograms in the R Project for Statistical Computing and similar statistical programs which still need such inputs.
Keywords: artemisia gemilini, artemisia bravifolia, genetic diversity, protein concentration, morphological characters, RAPD, R program, statistical analysis, protocol optimization
Family Asteraceae (Syn. Compositae, commonly known as sunflower family) is a large family comprising 13 subfamilies, 1620 genera and more than 23,600 species.1 Members of Asteraceae are found all over the world especially in tropical and subtropical regions including central and South America, central Asia, and Mediterranean areas. Some important genera of the family are Artemisia, Cichorium, Thymopsis, Zexmenia, Chrysanthellum Bradburia etc.2 Various species of genus Artemisia are used as ornamental, landscape reclamation, medicinal, antimicrobial, fumigant, forage crop and to make tea and liquor etc.3 Artemisia species are also used for treatment of malaria. Artemisin is a chemical which is derived from Artemisia annua and is widely used for the treatment of malaria.4,5
Taxonomically Artemisia is considered a complex genus because various closely related species have different morphological forms. In general, it is believed that previous classification of Artemisia is not certain. Previously genus Artemisia was subdivided into sub genera and sections. But with molecular biology based phylogenic studies, it is now more evident that previous sub division of the genus into sub genera and sections was not correct.6 Some important Artemisia species found in Pakistan include Artemisia absinthium L. Artemisia annua L. and Artemisia bravifolia Wall. ex DC., Artemisia gemilini, Artemisia dracunculus L.7‒9 Gilgit Baltistan has large number of medicinally important plant species including Artemisia gemilini and Artemisia bravifolia.
All over the world Artemisia species are being used by indigenous communities for stomachic, purgative, hysteria, asthma, skin diseases like scabies, malaria, ulcers, diabetes, high blood pressure, gastrointestinal ailments and selective killing of human breast cancer cells.6,10 All Artemisia species produce aromatic oils. Artemisia species are also important having many volatile molecules such as terpenes, phenol-derived aromatic and aliphatic components. Isolated a flavonid named “Eupatilin (5, 7-dihydroxy-3', 4', 6-trimethoxyflavone)” having anticancer and antioxidant properties to treat human melanoma (cell line A375). Antihelmintic agent “Santonin” had been isolated from Artemisia maritime. Many species of genus Artemisia have biotic and abiotic stress tolerance.10 In Pakistan especially in Gilgit Baltistan, Artemisia based folk medicines are used for treatment of various diseases including diabetes, ENT problems, gastric problems, intestinal worms, skin infections, thorax illness etc.9,11,12 Khan & Khatoon13 reported that in mountainous areas of Pakistan, whole Plant extract of Artemisia bravifolia is used for fever, cough, blood pressure and abdominal pains.
Giving due importance to medicinal values, much scientific work has been done in the world on various species of genus Artemisia. For example, Kumar et al.14 used Expressed Sequence Tag (EST) and Simple Sequence Repeat (SSR) primers to estimate the genetic diversity in Artemisia species from Himalaya region. Al-Rawashdeh15 and Badar et al.,16 used morphological characters and Randomly Amplified Polymorphic DNA (RAPD) primers to estimate genetic diversity in Artemisia species from Egypt. Pellicer et al.,17 used chloroplast DNA sequence to study phylogenetic relationship in Artemisia species. Unfortunately, in Pakistan not much work has been reported on various indigenous species of genus Artemisia. Objective of present research was to utilize recently developed DNA based techniques to study genomics/comparative genomics in Artemisia gemilini and Artemisia bravifolia (Figure 1).
Recently established province Gilgit Baltistan is the most magnificent mountainous area of the world surrounded by mighty mountain systems of Himalaya, Karakoram, Hindukush and Pamir. Latitude of the area is 35.3500°N, 75.9000°E. Plant material for the study was collected from 6 locations in GB viz., Low altitude Bunji (town of Bunji 1400 meter above sea level), medium altitude Bunji (3500 meters above sea level), high altitude Bunji (5500meters above sea level), Gilgit city (1500 meter above sea level), Bagrot valley (2700 meter above sea level), Yaseen valley, district Ghizer (1760 meter above sea level).
For biochemical analyses, leaf samples were used mainly because leaves of the Artemisia species are commonly used. Total crude protein was extracted using procedure described by Yeoh & Wong.18 Total crude Protein content was estimated using UV spectrophotometer. Relationship given by Grimsley & Pace19 that an absorbance of 1.3 (at 280nm wave length) is equal to 1 mg protein/ml was used.
Different tissues of plant material (leaves, seed and bark) were individually used to optimize extraction of DNA. Finally protocols originally described by Weining & Langridge20 and Doyl & Doyl21 were modified for successful isolation of total genomic DNA from bark of Artemisia species. Extraction was done using CTAB and 150 microliter beta marcaptoethanol in the DNA extraction buffer. The quality and quantity of the DNA was checked on 1% agarose/TBE gel. Amplifications of Artemisia DNA using 28 RAPD primers were performed using standard protocol.22
Basic Statistical analysis was carried out using computer program Past23,24 version 3.11. For genetic diversity analyses using SDS PAGE and RAPD primers, Genetic distances were estimated using following formula.25
Where GD, Genetic distance; dxy, number of common bands in two samples; dx, number of total fragments in sample number 1; dy, number of total fragments in sample number 2.
Artemisia gemilini was not found from medium and high altitudes Bunji (more than 3500 meter above sea level). The finding strengthened previous finding Khan & Khatoon.13 The two species were significantly different for eight morphological characters were studied viz; Plant height, number of branches, leaf length, leaf width, number of rachis on main stem, diameter of stem, diameter of branches and dry weight of plant. Mean values and basic statics for the morphological characters is presented in Table 1.
Plant height |
No of braches |
Leaf length (cm) |
Leaf width (cm) |
Stem diameter (cm) |
Diameter of branches(cm) |
No of rachis |
Dry weight (g) |
|||
A.G |
A.B |
A.G |
A.B |
A.G |
A.G |
A.G |
A.B |
A.G |
A.B |
|
Min |
80 |
30 |
19 |
31 |
5.14 |
2.58 |
2.5 |
0.26 |
8 |
12.27 |
Max |
122 |
62 |
50 |
76 |
8.14 |
4.78 |
8 |
4.6 |
9 |
458.76 |
Mean |
101.45 |
49.7 |
7.3 |
48.15 |
6.185 |
3.626 |
4.51 |
1.077 |
8.15 |
102.29 |
Median |
100.5 |
49.7 |
39 |
45.5 |
6.02 |
3.56 |
4 |
1.03 |
8 |
55.13 |
St error |
2.39 |
2.3148 |
1.8667 |
2.887 |
0.17 |
0.139 |
0.42 |
0.17 |
0.08 |
21.05 |
St |
10.71 |
10.352 |
8.3483 |
12.913 |
0.77 |
0.62 |
1.87 |
0.91 |
0.37 |
115.28 |
Coeff |
10.56 |
20.829 |
22.381 |
26.819 |
12.41 |
17.2 |
41.51 |
84.54 |
4.49 |
112.7 |
Table 1 Basic statistics for morphological characters studied in Artemisia gemilini and Artemisia bravifolia.
A.G, Artemisia gemilini; A.B, Artemisia bravifolia
Crude protein estimates in A. gemilini and A. bravifolia ranged from 17.6-20.7 % (Table 2). Crude protein content was highly significantly different in the 10 samples studied, t-value=54.97**, p-value=1.097E-12. In an earlier study, Rauzi26 reported 10-17 % crude protein content in leaves and flowers of Artemisia frigida.
S. no |
Sample |
Leaf crude protein % |
1 |
A. gemilini collected from low altitude Bunji (1400meter) |
19.2 |
2 |
A. bravifolia collected from low altitude Bunji (1400meter |
18.5 |
3 |
A. bravifolia collected from mid altitude Bunji (3500meter |
17.6 |
4 |
A. bravifolia collected from high altitude Bunji (5500meter) |
20.7 |
5 |
A. gemilini from Gilgit (1500meter) |
20.7 |
6 |
A. bravifolia from Gilgit (1500meter) |
20 |
7 |
A. gemilini from Bagrot (2700meter) |
19.2 |
8 |
A. bravifolia from Bagrot (2700meter) |
18.5 |
9 |
A. gemilini from Yaseen (1760meter) |
18.5 |
10 |
A. bravifolia from Yaseen (1760meter) |
20.7 |
Table 2 Estimation of crude protein concentration extracted from leaves of Artemisia gemilini and Artemisia bravifolia
t-value=54.97**, p-value =1.097E-12
Crude Protein isolated from leaves of Artemisia gemilini and Artemisia bravifolia samples was also studied using Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis (SD-PAGE) (Figure 2). Genetic distances calculated using bivariate data generated from SDS-PAGE results ranged from 0-75%. Protein band number 2, 3 and 4 were observed in Artemisia gemilini only. Protein band 2 and 4 were observed only in Artemisia gemilini collected from low altitude Bunji (1400 meter above sea level).
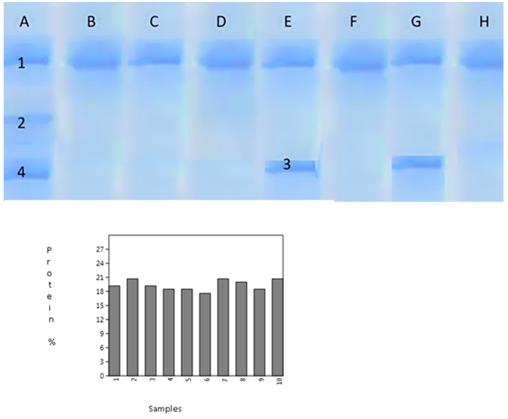
Figure 2 SDS-PAGE profile of 10 samples of genus Artemisia. Crude protein percentage graph. (A) Artemisia gemilini Low altitude Bunji, (B) Artemisia bravifolia Low altitude Bunji (C) Artemisia bravifolia Mid altitude Bunji (D)Artemisia bravifolia High altitude, Bunji (E) Artemisia gemilini Yaseen valley (F) Artemisia Bravifolia Yaseen valley (G) Artemisia gemiliniBagrot (H) Artemisia bravifoliaBagrot (I) Artemisia gemilini Gilgit (J) Artemisia bravifolia Gilgit.
An example of PCR amplifications of A. gemilini and A. bravifolia using RAPD primer GLA-03 is presented in Figure 3. Only reliably scoreable DNA fragments (DNA bands) were included in the analyses. DNA fragments were scored as present (1) and absent (0). A total of 125 DNA fragments were amplified using 28 RAPD primers giving an average of 4.5 DNA fragments per primer. Bivariate data matrix was used for the estimation of genetic diversity using UPGMA procedure described by Nei and Li (1979). Genetic distances calculate for all the 28 primers ranged from 0 to 100%. Three RAPD primers GL C-2, E7 and GL E-15 showed complete homozygosity (GD=0%) between Artemisia gemilini and Artemisia bravifolia. Eight RAPD primers viz; GL A-10, GL A-16, GL B-1, GL B-6, GL B-20, GL C-12, GL H-1 and GL I-8, showed 100% genetic diversity between Artemisia gemilini and Artemisia bravifolia (Table 3) (Table 4).
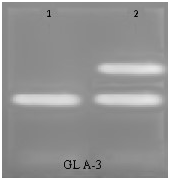
Figure 3 Amplification of A. gemilini (lane 1) and A.bravifolia (lane 2) DNA using RAPD primer GLA-03.
S.no |
Name of RAPD |
Genetic distance (%) |
1 |
GLA-03 |
50 |
2 |
GLA-10 |
100 |
3 |
GLA-14 |
50 |
4 |
GLA-16 |
100 |
5 |
GLA-18 |
67 |
6 |
GLB -01 |
100 |
7 |
GLB-05 |
50 |
8 |
GLB-06 |
100 |
9 |
GLB- 20 |
100 |
10 |
GLC-01 |
50 |
11 |
GLC-02 |
0 |
12 |
GLC-09 |
40 |
13 |
GLC-12 |
100 |
14 |
GLC-13 |
34 |
15 |
GLD-01 |
34 |
16 |
GLD-03 |
40 |
17 |
GLD-09 |
67 |
18 |
GLE-05 |
50 |
19 |
GLE-07 |
0 |
20 |
GLE-15 |
0 |
21 |
GLF-03 |
34 |
22 |
GLF-10 |
10 |
23 |
GLG-02 |
67 |
24 |
GLG-16 |
34 |
25 |
GLH-01 |
100 |
26 |
GLI-01 |
25 |
27 |
GLI-08 |
100 |
28 |
GLK-08 |
25 |
Table 3 Genetic distances among Artemisia gemilini and Artemisia bravifolia using 28 RAPD
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
|
2 |
70% |
||||||||
3 |
70% |
0% |
|||||||
4 |
70% |
0% |
0% |
||||||
5 |
75% |
50% |
50% |
50% |
|||||
6 |
70% |
0% |
0% |
50% |
50% |
||||
7 |
75% |
50% |
50% |
0% |
0% |
50% |
|||
8 |
70% |
0% |
0% |
50% |
50% |
0% |
50% |
||
9 |
75% |
50% |
50% |
0% |
0% |
50% |
0% |
50% |
|
10 |
70% |
0% |
0% |
50% |
50% |
0% |
50% |
0% |
50% |
Table 4 Genetic distances (G.D) in 10 samples of genus Artemisia based on crude leaf protein
1=Artemisia gemilini Low altitude Bunji , 2= Artemisia bravifolia Low altitude Bunji 3= Artemisia bravifolia Mid altitude Bunji 4=Artemisia bravifolia High altitude, Bunji 5= Artemisia gemilini Yaseen valley 6= Artemisia Bravifolia Yaseen valley 7= Artemisia gemilini Bagrot 8 = Artemisia bravifolia Bagrot 9= Artemisia gemilini Gilgit 10= Artemisia bravifolia Gilgit.
Taxonomically Artemisia is considered a complex genus because various closely related species have different morphological forms. In general, it is believed that previous classification of Artemisia (based on morphological characterization) is not certain. Using molecular biology based phylogenic studies; it is now more evident that previous sub division of the genus into sub genera and sections was not correct.6 Artemisia gemilini (local known as “Khakhamus” in Shina language) is a perennial species, usually bitter-aromatic, glabrous, hairy herb with tap root. It is -more than 1.0meter in height. Artemisia bravifolia (Locally known as “Zoon”) is a woody shrublet, up to 70cm tall; greenish to brownish-yellow, rarely reddish vegetative and flowering stems which is upright, thick, much-branched and woody.17,23 These two medicinally important species from mountainous areas of GB were characterized using morphological, biochemical and DNA based markers.
Plant material for the study was collected from a wide range of areas in the province of Gilgit Baltistan. Samples were collected from 6 locations. Major emphasis was given to Bunji area, district Astore but additional samples were also collected from districts Gilgit and Ghizer. Plant samples were collected from various altitudes ranging from 1400 meter above sea level to 5500meters above sea level. Average values for various morphological characters observed during present research were similar to those reported earlier.17 Highly significant differences among the two species indicated presence of sufficient amount of genetic variation within the genus. Lea protein content was similar to those reported by Rauzi.26
Extra protein bands observed in Artemisia gemilini collected from high altitude. These protein fragments should be further studied in detail and can be used to strengthen our knowledge regarding taxonomic classification and genetic structure of genus Artemisia. Total genomic DNA isolation from bark technique was optimized which can be used at larger scale under locally available conditions (equipment and chemicals etc). Twenty-eight RAPD primers were used for estimation of genetic distances between the two species. Species specific DNA fragments amplified using RAPD primers GLA-10, GLA-16, GLB-1, GLB-6, GLB-20, GLC-12, GLH-1 and GLI-8 should be sequenced and may be used for synthesis of larger species specific RCR primers. It was concluded that enough genetic variability exists in genus Artemisia in Gilgit Baltistan. This genetic variability can be utilized for the improvement of Artemisia gemilini and Artemisia bravifolia. As taxonomically Artemisia is considered a complex genus because various closely related species have different morphological forms.6 It is suggested that DNA markers should be used to solve ambiguities in taxonomic classification of genus Artemisia. In addition, it is also recommended that species specific protein bands (band number 2, 3 and 4) should be studied in more detail.
Authors declare that there is no conflict of interest.

©2019 Gul, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.