Advances in
eISSN: 2373-6402


Research Article Volume 8 Issue 3
Department of Biotechnology, Kakatiya University, Plant Biotechnology Research Laboratory, India
Correspondence: Rama Swamy Nanna, Plant Biotechnology Research Laboratory, Department of Biotechnology, Kakatiya University, Warangal-506009, Telangana, India
Received: October 24, 2017 | Published: June 4, 2018
Citation: Marka R, Nanna RS. Optimization of factors affecting agro bacterium-mediated genetic transformation in groundnut ( arachis hypogaea l.). Adv Plants Agric Res. 2018;8(3):275-282. DOI: 10.15406/apar.2018.08.00327
The efficiency of Agrobacterium-mediated genetic transformation of groundnut is dependent on various factors such as type and age of explants, antibiotics used for selection, Acetosyringone (AS), duration of co cultivation and infection period. Hence, we have attempted to standardize these parameters to improve the Agrobacterium mediated genetic transformation efficiency in groundnut cv ICG 13942. To evaluate the optimization of factors affecting Agrobacterium mediated genetic transformation in which we used three different explants (Deembryonated cotyledon-DC, Leaflet-LL, Cotyledonary node-CN) and studied, the effect of inoculum concentration (0.5, 1.0 and 1.5 OD600 of A. tumefaciens), pre culture period (1-11 days), co cultivation period (1-7 days), Acetosyringone concentration (25-200mg/L) and antibiotic (Kan) concentration (25-150mg/L) on the efficiency of genetic transformation. The transformation was verified by GUS staining and by means of PCR amplification of the uidA and nptII genes. The optimized factors for A. tumefaciens LBA 4404 harboring pBAL2 binary vector mediated transformation in groundnut cv ICG 13942 are: Pre culture period-3 days, Concentration of A. tumefaciens-1.0 at OD600, Co-cultivation period-4 days, Kan concentration-100mg/L for DC &CN, 75mg/L for LL explants, Concentration of AS-100mg/L during infection and co cultivation and Cefotaxime concentration-250mg/L. Thus, the optimized protocol showed the enhanced transient transformation frequencies such as 18.13% for DC, 15.33% for LL and 12.32% for CN explants.
Keywords: agrobacterium tumefaciens, arachis hypogaea, GUS expression, npt-II, polymerase chain reaction
KAN, kanamycin; AS, acetosyringone; CN, cotyledonary node; LL, leaflet; DC, de emryonated cotyledon; nptII, neomycin phosphotransferase; NAA, α-naphthalene acetic acid; BAP, 6-Benzylaminopurin
Groundnut/Peanut (Arachis hypogaea L.) has great potential in third world countries to reduce hunger and malnutrition.1 It is widely grown in the semi-arid tropics and India ranks second by 12.5% in world production.2 The production of groundnut is constrained by different biotic and abiotic factors by causing decrease in yield.3 To develop stress tolerance through conventional breeding takes longer time and passes through various incompatibility barriers. To overcome these, the biotechnological tools play a great role for developing stress tolerance through introduction of alien genes into elite germplasm of crop plants through genetic transformation technology.4-6 The various factors that affect significant differences in T-DNA delivery and transformation included explant type, duration of pre-culture of ex plant, incubation period in bacterial suspension, the concentration of A. tumefaciens during infection, the use and concentration of inducers such as Acetosyringone during infection, antibiotics used for selection and the extent of the co-culture period were optimized to establish a transformation protocol of common bean (Phaseolus vulgaris).7 and in chickpea.8 Legumes in general are recalcitrant to tissue culture and Agrobacterium mediated transformation is also highly genotype specific.9 Several reports have been published on efficient regeneration from diverse explants of peanut,10,11 but less success has been recorded with genetic transformation of A.hypogaea. This is due to lack of efficient protocols to induce adventitious shoot buds after the transformation.12-14 Recently we have developed the reproducible regeneration protocol in groundnut cv ICG 13942.15-17 The aim of this work was to develop a reproducible and efficient method of A. tumefaciens mediated genetic transformation and to evaluate the factors that affect the genetic transformation efficiency by using marker genes (nptII; uidA) and later the same was used for Agrobacterium mediated transformation with the Tc Chitinase-I gene to develop the resistance to Tikka disease in cv ICG 13942 for the first time. Therefore, in the present report we describe the successful transformation and regeneration of putatively transformed plants in groundnut cv ICG 13942.
Plant material, culture initiation and maintenance
The mature seeds of groundnut cv ICG 13942, obtained from the germplasm bank of ICRISAT, Patancheru, Hyderabad, Telangana, India were used. Dried seeds were washed under running tap water for 10-15 min followed by treating with Tween-20 (5%-v/v) for 5 min. Later these were rinsed twice in sterile distilled water. Afterwards the seeds were surface sterilized with 0.1% (w/v) HgCl2 for 8 min followed by rinsing in sterilized distilled water for 3-4 times under aseptic conditions and soaked for 24 h in sterile distilled water. The experiments were conducted on modified MS18 medium (MMS medium) containing MS basal salts and B5 vitamins,19 100 mg/L myo-inositol and 30 g/L sucrose. The pH of the medium was adjusted to 5.8±0.02 with either 0.1N NaOH or 0.1N HCl before addition of agar. The medium was solidified with 0.8% Difcobacto-agar and autoclaved at 121ºC under 15 psi for 15-20 min. All the cultures were incubated at 25±2ºC under cool white fluorescent light (1600lux) with a 16/8 h (light/dark) photoperiod.
Explants preparation
Three different kinds of explants i.e. de-emryonated cotyledon (DC), leaf let (LL) and cotyledonary node (CN) were used for transformation. Mature dry seeds were imbibed in sterile distilled water for 24 h after surface sterilization and kept for germination on autoclaved filter paper bridge/boat in ½ strength liquid MMS medium in culture tubes. In the case of CN explants, they were excised from 7days old in vitro grown seedlings and removed the cotyledons, radical, shoot tip parts, LL explants from axenic seedlings (2 weeks old) were cut into 0.8-1.0 cm2 size and DC explants were isolated from the soaked (24 hrs) seeds and these were inoculated on the following medium: CN explants on MMS+20 mg/L BAP (SIM),15 DC explants on MMS+0.5mg/L IAA+15 mg/L TDZ (SIM)16 and Leaflet explants on MMS+2mg/L AgNO3+10mg/L BAP+0.5mg/L IAA (SIM).17
Agrobacterium strain and plasmid vector
The Agrobacterium tumefaciens strain LBA 4404 harboring the binary vector pBAL2 (18.8Kb) was used for optimization of transformation protocol in groundnut cv13942. The binary vector pBAL2 consists of reporter gene (β-glucuronidase-gus/uidA) by the CaMV 35S promoter and terminator sequences and the selectable marker gene (npt II) driven by the nopaline synthase (NOS) promoter and terminator sequences respectively (Figure 1).
Transformation and regeneration of transformants
The A.tumefaciens strain LBA4404 harboring pBAL2 was grown on YEM agar medium containing 50mg/L of Kan and 10mg/L Rif. A single bacterial colony was inoculated in 50ml liquid YEM medium containing the same antibiotics and grown overnight at 28ºC in an incubator shaker at 120rpm. 5ml of this overnight culture was re inoculated in 50 ml fresh YEM liquid medium containing 50mg/L Kan and grown overnight. The bacterial cell density was determined at OD600 of 0.5 to 1.5. Later the bacterial suspension was pelleted at 5,000rpm for 10 min and re suspended in 25ml hormone-free liquid half-strength MS medium. The explants, DC, LL and CN were pre-cultured for 1-11 days on shoot induction medium (SIM) prior to infection with bacteria. The trimmed explants were immersed in Agrobacterium suspension containing 25-200mg/L AS for 10-30 minutes and subjected to manual shaking. Later, the explants were dried on sterile tissue paper and co-cultivated for 1-7 days on SIM along with 100mg/L AS and incubated in the dark at 28±2ºC. Afterwards the explants were washed with 250mg/l cefotaxime for 5 min followed by rinsing with sterile distilled water thrice for 5 min and dried on sterile tissue paper. All these explants were cultured on selection medium containing SIM+75mg/L Kan for leaflet and 100mg/L Kan for DC and CN explants and 250mg/l cefotaxime for 2-3 weeks. The explants with KanR shoots were sub cultured on SIM with 50mg/L Kan for further proliferation of shoots. Later each micro-shoot was excised and cultured on MMS+50 mg/L Kan+0.5 mg/L BAP for shoot elongation. After elongation, the micro-shoots were transferred on to root induction medium (RIM) containing 1mg/L NAA+50mg/L Kan. All the cultures were incubated at 25±2 ºC with a 16/8 hr photoperiod. These KanR plantlets were shifted to plastic cups containing the soil mix, sand: soil: vermiculite (1:1:1) and acclimatized in the growth chamber. Established plantlets were transferred to green house and allowed to grow in an earthenware pots for maturity.
Histochemical GUS analysis
The commercial kit “β-Glucuronidase Reporter Gene Staining Kit” was performed, following the protocol recommended by the supplier (Sigma Aldrich), after which it was incubated at 37°C for 24h. The shoots were then rinsed twice with ethanol at 70% and once with acetone at 50%, according to the recommendations of Jefferson et al.20 Finally, the number of explants presenting blue coloring and their percentage per treatment was registered by stereoscopic observation.
PCR analysis of putative transformants
The leaf genomic DNA from T0 plants of cv ICG 13942 was isolated by Cetyl Trimethyl Ammonium Bromide (CTAB) method and used for molecular characterization of putative transgenic by PCR using nptII and uidA gene specific primers. The npt II primer sequences were, (F) 5’-GCT TGG GTG GAG AGG GCT ATT-3’ (R) 5’-AGA ACT CGT CAA GAA GGC GA -3’ and uidA gene specific primer sequences were, (F) 5′- TTT AAC TAT GCC GGG ATC CAT CGC - 3′ (R) 5′- CCA GTC GAG CAT CTC TTC AGC GT - 3′. The PCR for nptII was carried out by initial denaturation at 94°C for 5 min followed by 35 cycles of 94°C for 1 min, 54°C for 1 min and 72°C for 1.30 min and final extension at 72°C for 10 min. A PCR programme for uidA gene was carried out by initial denaturation at 94°C for 5 min followed by 35 cycles of 94°C for 1 min, 64°C for 1.30 min and 72°C for 2 min and final extension at 72°C for 10 min. Plants confirming positive with PCR were taken to maturity and their seeds were collected. The genomic DNA from the untransformed control plants and PCR +ve plants for nptII and uid A genes were used as negative and positive controls, respectively. The amplified fragments were separated by electrophoresis on a 0.8% agarose gel, using TE buffer. Gels were stained with ethidium bromide, photographed by using gel documentation system (Biorad).
Data analysis
The transformation efficiency was calculated in different explants.
The effect of different parameters on the transformation was carried out. The parameters include pre culture of explants, bacterial cell density, bacterial infection period, co cultivation period, concentration of AS and cefotoxime. Hence, all the experiments were carried out with Agrobacterium tumefaciens strain LBA 4404 harboring the binary plasmid pBAL2 contains nptII and uidA marker genes and co cultivation on MMS medium supplemented with appropriate PGRs (SIM) with different explants (DC, LL and CN) of groundnut cv ICG 13942.
Factors affecting genetic transformation
Effect of different OD at 600nm
The pre cultured (3 days) DC, LL and CN explants were submerged in bacterial suspension at different OD600 values of 0.5, 1.0, 1.5 bacterial suspension for 20 min and were cultured on SIM containing 100 mg/L AS for 4 days. The bacterial concentration of 1.0 value at OD600 resulted the highest percentage of regeneration with transformation efficiency in DC (82.60%), LL (74.25%), CN (61.13%) explants of cv ICG 13942 (Figure 3a).

Figure 2a-i Determination of antibiotics sensitivity (Kan) in three different types of explants (DC, CN, LL) of groundnut cv ICG 13942. a-c) Shoot buds proliferation on SIM without antibiotics (control), Shoot buds formation at 100 mg/L Kan, Inhibition of shoot buds development at 125mg/L Kan (Note the necrosis of explants) in DC explants respectively, d-f) Shoot buds proliferation on SIM without antibiotics (control), Shoot buds formation at 100 mg/L Kan, Inhibition of shoot buds development at 125mg/L Kan (Note the necrosis of explants) in CN explants respectively, g-i) Shoot buds proliferation on SIM without antibiotics (control), Shoot buds formation at 75 mg/L Kan, Inhibition of shoot buds development at 100 mg/L Kan (Note the necrosis of explants) in LL explants respectively.
Effect of pre-culture period (days)
To test whether the pre culture period has influence on transformation, the DC, LL and CN explants were cultured on SIM for 1-11 days before the selection on Kan containing medium and also prior to the infection with Agrobacterium strain. Of these, the pre cultured explants for three days on SIM have shown the maximum percentage of response in all the three explants of peanut cv used (Figure 3b). Gradually the percentage of transformation was found to be reduced after 3rd day of pre culture because the explants pre cultured for 5th, 7th, 9th and 11th days were unable to receive the Agrobacterium due to the excessive growth of the explant. Among the explants, the DC (69.11%) and LL (66.23%) explants have shown the maximum transformation efficiency followed by CN (64.92%) explants of cv ICG 13942.
Effect of co-cultivation period
The pre cultured (3 days) DC, LL and CN explants were submerged in bacterial suspension for 20 min and were co cultivated on SIM containing 100 mg/L AS for 1-7 days. Four days of co-cultivation resulted the highest percentage of regeneration with transformation efficiency in DC (66.22%), LL (60.14%), CN (57.23%) explants of cv ICG 13942 (Figure 3c).
Effect of infection period
The pre cultured explants viz., DC, LL and CN were submerged in bacterial suspension for 10, 15, 20, 30 min, and they were co cultivated on SIM containing 100 mg/L AS for 4 days. 20 min infection period had shown the highest percentage of transformation efficiency in DC (58.13%), LL (52.20%), CN (48.62%) explants of cv ICG 13942 (Figure 3d). For more than 20 min infection period, led to the explant abortion caused by bacterial contamination by reducing the transformation frequency.
Effect of antibiotics sensitivity (Kan)
The pre cultured DC, LL and CN explants were inoculated on SIM with different concentrations (25-150mg/L) of Kan (Figure 3e). The percentage of shoot induction response along with multiple shoots development/explants was found to be reduced as the concentration of Kan increased in all the explants tested. More than 90% of shoot induction response was inhibited at 100mg/L Kan in DC, CN explants (Figure 2a–f) and more than 85% at 75mg/L Kan in LL explants (Figure 2g–i). Beyond that level of Kan, showed the bleaching and necrosis of the explants.
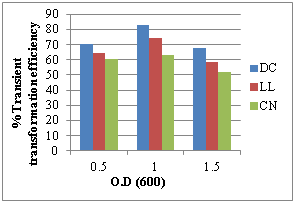

a) Effect of different values of O.D at 600nm b) Effect of pre-culture period

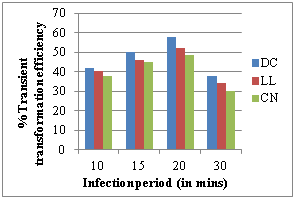
c) Effect of cocultivation period d) Effect of infection period
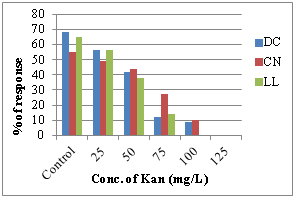
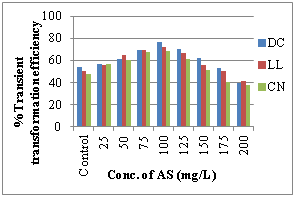
e) Effect of antibiotics sensitivity f) Effect of Acetosyringone concentration
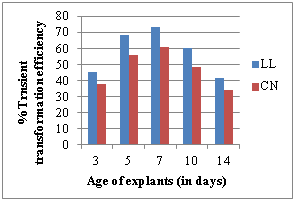
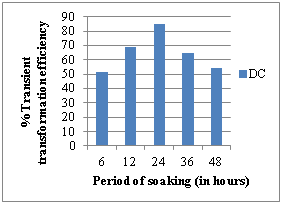
g) Effect of age of explant h) Effect of soaking period

i) Effect of explant type
Figure 3a-i Transient transformation efficiency with different parameters.
Effect of Acetosyringone (AS)
Different levels of AS (25- 200 mg/L) was tested and at 100 mg/L AS was found to be the best for enhancing the transformation efficiency in groundnut cv ICG 13942. The maximum percentage (76.54%) of GUS positive explants was observed in DC explants, followed by 71.78% in LL and 68.21% in CN explants (Figure 3f).
Effect of explant age
Seven days old seedlings and 24 hours soaked seeds exhibited more GUS expression by showing maximum percentage of transformation efficiency in all the explants used. Transformation efficiency was thus affected by the age of the explant source with optimal transformation efficiencies of 85.13%, 73.13% and 61.09% for DC, LL and CN explants respectively (Figure 3g-h).
Effect of explant type
Susceptibility to Agrobacterium was dependent on the type of explant. In order to improve the protocol of genetic transformation, LL, CN explants were excised from 7-days old axenic seedlings and DC explants were excised from 24 hours soaked seeds and compared for their transformation efficiencies. The difference in transformation efficiencies was recorded. The highest transformation frequency was observed for DC (18.13%) followed by LL (15.33%) and CN (12.32%) explants (Figure 3i).
Effect of cefotaxime concentration
After four days of co-cultivation, the explants (DC, LL & CN) were washed with sterile half-strength MS liquid medium containing 250mg/l Cefotaxime to prevent the over growth of Agrobacterium in the infected explants. Subsequently these infected explants were cultured on the selection medium containing SIM+100mg/L Kan+250mg/L Cefotaxime for DC & CN, SIM+75mg/L Kan+250mg/L Cefotaxime for LL explants for 2 weeks. Cefotaxime totally arrested the overgrowth of Agrobacterium which causes the death of the explants.
Confirmation of transformants
All these standardized parameters were used for A. tumefaciens LBA 4404 mediated transformation in groundnut cv ICG 13942. After co cultivation for 4 days, the DC, LL and CN were cultured on their respective selection medium for 2-3 weeks. The KanR shoots were developed from the transformed cells (Figure 4a–c). The explants with KanR shoots were sub cultured on SIM+50mg/L Kan for further proliferation. The individual micro-shoot was elongated on MMS+50mg/L Kan+0.5mg/L BAP. These elongated micro-shoots were rooted on RIM+1.0mg/L NAA+50 mg/L Kan. The KanR plantlets (T0) were successfully acclimatized and maintained in the green house (Figure 4i).

Figure 4a-i Agrobacterium mediated transformation and GUS Assay of transgenic shoots (KanR) developed from DC, CN and LL explants of cv ICG 13942 by using the binary vectorpBAL2. a-c. a-c. Induction of KanR shoots on selection medium after 4 weeks of incubation from DC, CN and LL respectively, d) In vitro rooting of shoots on RIM supplemented with 1.0 mg/L NAA+ 50mg/L Kan (Note profuse rhizogenesis), e) Non transformed shoots (control-WT), f-h) Transgenic shoots developed from DC, LL and CN explants respectively, i) Plants are shifted to plastic pots containing soil mix and maintained in the green house.
GUS assay
To compare transient and stable T-DNA transformation, GUS analysis was done in explants immediately after co-cultivation as well as in young growing tissues of T0 generation of putative transformants. Transient GUS expression was not consistent, however, stable GUS expression was observed in young T0 shoots of DC, LL and CN explants (Figure 4F–H). No GUS activity was observed in non-transformed (control) explants (Figure 4E). Explants without pre culture and mechanical injuries, at the site of regeneration, upon co cultivation with Agrobacterium showed no staining (DC, LL and CN). Whereas, pre cultured explants with mechanical injuries showed GUS activity at the wounded regions of the explants. Thus, we have found the typical expression pattern of the CaMV 35S promoter regulated uidA gene in transformed tissues. These results show the presence of GUS expression only in transformed tissues, clearly demonstrate the stability of the integrated gene in the putatively transformed plants of cv 13942 of groundnut.
Molecular confirmation
In the present study, molecular analysis was carried out by using PCR technique in T0 plants. The amplification of the presence of nptII and uidA (gus) genes in KanR putatively transformed plants (T0) was confirmed. These putatively transformed plants were obtained after three cycles of culture on selection medium after co cultivation with A. tumefaciens LBA 4404 strain harboring the binary vector pBAL2. The gene specific primers for nptII and uid ‘A’ genes were used for PCR amplification and the amplified DNA fragments were found 750bp and 1.9 Kb respectively (Figure 6) (Figure7). The transformation efficiency was estimated on the basis of PCR +ve plants (NPT-II and GUS) developed in the present investigations. Out of 915 KanR plants developed only 141 plants showed the PCR +ve (nptII & gus) with 15.40% of transformation efficiency in three different types of explants (DC, LL and CN) used in cv ICG 13942 (Figure 5). Thus, the presence of the uidA and nptII genes in the putatively transformed plants (T0) of groundnut cvs was confirmed by PCR analysis. PCR analysis revealed the presence of 1.9 kb uidA and 750bp nptII DNA fragments amplified from the genomic DNA of all T0 transgenic plants. However, PCR products of uidA and nptII genes were not seen in untransformed (control - WT) plants (Figures 6,7), Lane C). All the samples of transgenic plants of cv 13942 gave the predicted size of DNA fragment of uidA gene (1.9Kb) (Figure 6, Lane 1-7). The band was not detected in the DNA sample from an untransformed (control) plant (Figure 6, Lane C). The DNA product with the size of 750bp nptII gene was amplified from total genomic DNA of the T0 transgenic plants of cv 13942 of groundnut (Figure 7, Lane 1-7). This DNA fragment was not detected in the untransformed (control-WT) plants (Figure 7, Lane C). As a positive control, the nptII gene product was also amplified from the pBAL2 plasmid (Figure 7), Lane P). Based on the development of KanR plants, GUS assay and PCR detection, the presence of both uidA and nptII genes were confirmed in the T0 transgenic groundnut plants.
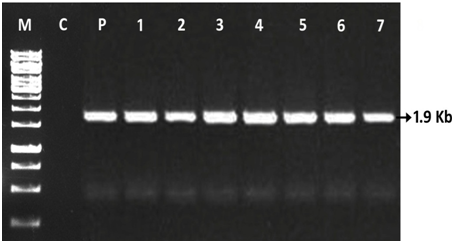
Figure 6 PCR screening of T0 plants for the presence of the uidA gene using gene-specific primers in cv ICG 13942 of groundnut. Lane M, DNA marker; Lane C, Control plant DNA (-ve control-WT); Lane P, Plasmid pBAL2 (+ve control); Lanes 1-3, T0 plants developed from DC; Lanes 4-5, T0 plants developed from LL; Lanes 6-7, T0 plants developed from CN explants.
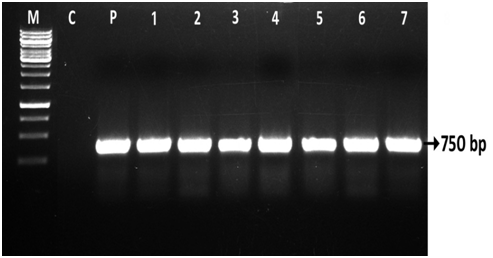
Figure 7 PCR screening of T0 plants for the presence of the nptII gene using gene-specific primers in cv ICG 13942 of groundnut. Lane M,DNA Marker; Lane C, Control plant DNA (-ve control-WT); Lane P, Plasmid pBAL2 (+ve control); Lanes 1-3, T0 transgenic plants developed from DC; Lanes 4-5, T0 transgenic plants developed from LL; Lanes 6-7, T0 transgenic plants developed from CN explants.
During the present study, we have attempted for optimization of factors affecting Agrobacterium mediated genetic transformation from individual explant of DC, LL, CN and found best transformation response in DC (18.13% Transient transformation efficiency) of groundnut cv. ICG 13942. There are various parameters which affect transformation efficiency as explored in groundnut.21 The highest transient transformation efficiency was observed for DC (18.13%) followed by LL (15.33%) and CN (12.32%) explants (Figure 3I) in the present investigation. Thus, the transformation efficiency is dependent upon the type of explant in groundnut and similar observations were also made in this crop: 2%,22, 16.5%23 when DC was used as explants, 2.2%,23 11.3%23 when LL was used as explants and 5.2%,24 8.5% 24 when used as CN explants.
Thus, we have achieved the increased level of transformation efficiencies in DC, LL and CN explants in comparison to earlier reports. The density of Agrobacterium suspension plays an important parameter. In the case of groundnut cv ICG 13942, the O.D600 1.0 gave maximum transient transformation efficiency 82.60% in DC, 74.25% in LL and 63.23% in CN explants while an O.D of 1.5 resulted in decreased transient transformation efficiency 67.43 % in DC, 58.63% in LL and 52.12% in CN explants (Figure 3A). Higher bacterial density in the medium may lead to severe necrosis of tissue.25 Pre-culture is an important parameter for genetic transformation of plants because in this step, to reduce the wound stress and enhance the attachment of bacterial cells at the wound site by increasing the entry of T-DNA part into the host cells. Present study showed that, the Transient transformation efficiency (T.T.E) was high 69.11% in DC, 66.22% in LL, 64.92 in CN explants after 3 days of pre-culture period while it decreased after 5-11 days of pre culture. Similarly, the importance of pre-culture during transformation studies was also reported in peanut.26 Co-cultivation is another important parameter for genetic transformation of plants because in this step, T-DNA is incorporated into plant genomic DNA.27 Our results showed that, the T.T.E was high 62.22% in DC, 60.14% in LL and 57.23 in CN after 4 days while it decreased after 5-7 days of co cultivation (Figure 3c). Co-cultivation for 4 days was shown to be acceptable in groundnut for better transformation while the longer for co-cultivation does not help to increase transformation efficiency in different legumes.28 It is reported that, secretion of Acetosyringone during wounding in plant cells induces the transcription of the virulence genes of Agrobacterium. Addition of Acetosyringone can improve transformation efficiency, in various monocots and some dicot plants which fail to secrete Acetosyringone.27
However in our study, addition of 100 mg/L Acetosyringone showed that, the T.T.E was high at 76.54% in DC, 71.78% in LL, 68.21% in CN while decreased at 125-200mg/L. According to our observations 20 min infection period had shown the highest percentage of transformation efficiency DC (58.13%), LL (52.20%) CN (48.62%) while it decreased after 30 min (Figure 3d). An extension of infection period for more than 20 min, led to the explant abortion caused by bacterial contamination which reduced the transformation frequency.29 Agrobacterium tumefaciens harboring pBAL2 plasmid carrying nptII and uidA genes were used for genetic transformation of groundnut. The present instigation showed more than 90% of shoot induction response inhibition at 100 mg/L Kan in DC, CN explants and more than 85% at 75 mg/L Kan in LL explants (Figure 3e). Beyond that level of Kan, showed the bleaching and necrosis of the explants. Similar findings were also reported in chickpea.30 For the effect of cefotaxime 250mg/L shows the inhibiting effect of Agrobacterium in DC, LL and CN in groundnut cv ICG 13942. Coherent to our results, in the case of Jatropha curcas, 250mg/L Cefotaxime supported the callus induction and shoot regeneration in cotyledon explants and therefore, cefotaxime was used to inhibit Agrobacterium growth after co-cultivation with cotyledon explants.31,32 In castor also similar results were found that overgrowth of Agrobacterium was controlled by 250mg/l Cefotaxime during transformation of embryo axes and shoot apices.33 According to our literature survey, this is the first report of Agrobacterium mediated genetic transformation of groundnut cv ICG 13942. Thus, the protocol developed during the present investigation can be used for genetic transformation experiments to introduce any novel gene into peanut.
We are grateful to University Grants Commission, New Delhi for financial assistance as Fellow under UGC-BSR–RFSMS (F.4-1/2006(BSR)/7-211/2009(BSR) dt.26-02-2013.). We thank Dr. HD. Upadhyaya ICRISAT, Patancheru, Hyderabad (TS), and India for the germplasm (seeds) of cv ICG 13942 and Prof. N. Jayabalan Department of Plant Science, Bharatidasan University, Thiruchurappally, Tamil Nadu, India, for the gift of pBAL2.
The author declares there is no conflict of interest.

©2018 Marka, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.