Advances in
eISSN: 2373-6402


Research Article Volume 8 Issue 2
1Department of Agricultural Science, Universiti Pendidikan Sultan Idris, Malaysia
2Department of Crop Science, Faculty of Agriculture and Food Science, Universiti Putra Malaysia, Malaysia
3Department of Crop Science, Faculty of Agriculture, Universiti Putra Malaysia, Malaysia
Correspondence: Norhanizan Usaizan, Department of Agricultural Science, Universiti Pendidikan Sultan Idris, 35900 Muallim Perak, Malaysia
Received: February 01, 2018 | Published: March 23, 2018
Citation: Usaizan N, Abdullah NAP, Saleh G, et al. Genetic diversity o f physalis minima L. accessions based on morphological traits. Adv Plants Agric Res. 2018;8(2):151-157. DOI: 10.15406/apar.2018.08.00306
Physalis minima L. is a member of Solanaceae family. This plant is commonly found in disturbed area and known as weed in Malaysia. P. minima contains high amount of phenolic, alkaloids and physalin compounds, indicating its potential as medicinal plant. Studies were conducted to analyze diversity and relationship within and among populations based on morphological characteristics. Nineteen populations were collected from 11 states of Peninsular Malaysia. The genetic diversity of germplasm collected was estimated using 16 qualitative and quantitative morphological characteristics. Results showed that the 19 populations of P. minima possessed identical qualitative characteristics. However, significant variations were detected for all quantitative traits measured among the 19 P. minima populations using ANOVA. Based on the variation among quantitative traits, the 19 populations were grouped into five clusters using UPGMA clustering method. This indicates the existence of different heterotic group among these populations. Crosses among populations from different heterotic group can cause high heterosis for important traits in P. minima. This information can be utilized in future breeding program to improve local P. minima varieties in Malaysia.
Keywords: Physalis, Solanaceae, ISSR, genetic diversity
Physalis minima or locally known in Malaysia as pokok letup-letup is a diploid (2n=24) plant and is one of the very important species in Solanaceae family Olmstead et al.1 It is native to warm temperate and subtropical region throughout the world. Physalis is a genus of 80 to 100 species of mostly neotropical herbs. The plant is an annual or short- lived perennial with less hairy as compared to a few other species. Physalis minima can grow well in most soil types particularly sandy soil. This plant commonly found as weeds in Malaysia Azlan et al.2 It has broad leaves and grows rapidly on disturbed area which makes it difficult to control. Whole part of the plant can be used as a treatment for ulcer and the decoction of the whole plant have the potential to decrease cancer cell activity Zakaria & Mohamad3 Extracts of the stem, leaf and unripe fruit contain photochemical substances such as flavonoids, terpenoids, vitamins, alkaloids and antibacterial substance which make it valuable as a medicinal plant Nathiya & Dorcus.4 The most interesting feature of this plant is the fruits. The yellowish edible berry is encapsulated by the calyx and it is a popular seasonal fruit in Europe. The fruit has been used as decoration in culinary, ingredient for delicious salads, desserts and also as flavoring in jams and jellies. Recently, the demand for this fruit has increased due to its potential as antioxidant and anticancer Pietro et al.;5 Azlan et al.;2 Shariff et al.6 The fruit has also reported to contain high amount of vitamin A and C and has the potential to be an alternative source for ascorbic acid for world consumption El-Sheikha et al.7 Morphological descriptions have traditionally been the means by which scientists investigate population structure and their genetic diversity. Mendel used morphological characteristics to develop his understanding of genetics and inheritance, while Darwin observed various morphological characteristics in domesticated plants and animals to develop his theory of natural selection and evolution. With recent revolutionary advances in molecular methods to characterize genetic diversity, phenotypic and molecular evaluations are now complementary practices Sattler & Rutishauser.8 Before the advent of molecular markers, morphological characterizations were the obligatory method for assessing genetic variability and systematize populations. Although morphological characterization provides only an indirect method to measure genetic variability, there are many documented studies that have used it successfully as a tool for classification of plants. Even today, morphology remains useful because germplasm curators and plant breeders usually routinely record morphological data during germplasm regeneration or evaluation Hokanson et al.9 Comparing phenotypic characteristics across populations is a relatively simple method for estimating variation and providing basic assessments of inter- and intra- population differences Parsons & Hermanutz.10 In Malaysia, Physalis minima is classified as a weeds and the numbers of Physalis species or varieties are relatively unknown. Weeds growing in cultivated areas are usually characterized as having high phenotype plasticity and genetic adaptability. They are frequently well adapted to disturbance and seed prolifically Adahtl.11 In addition, the colonization species, including exotic weeds such as Physalis minima will have low genetic variability since they have been through a genetic bottleneck effect after their introduction. However, the genetic variation of Physalis minima in Malaysia has not been reported. In this experiment, Physalis minima populations were collected from all states in Peninsular Malaysia. Qualitative characteristics of the plants were evaluated for their performance. The results were used to reveal phenotypic and genetic correlations of the traits measured, and to investigate the relationships among the populations based on their agronomic performance.
Germplasm collection
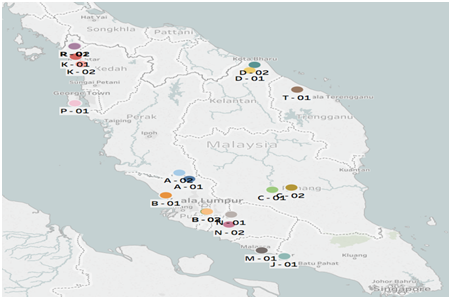
Hundred and thirty individual plants samples of Physalis minima were collected from 11 states of peninsular Malaysia from April 2011 to December 2012 (Table 1) (Figure 1). The sites of collections included farm, plantation area (especially of oil palm (Elaeis guineensis)) and the road site bush. The exact sampling sites with the local site names and code are as indicated in Table 1. Germplasm collection and sampling were done according to method of Hawkes12 where sampling was made every 30 km following state roads in different directions. From each site five to ten plants at the stage of flowering and fruiting were taken. At least 10 samples were collected from every state.
Morphological study
One hundred and thirty fresh samples from 19 populations were analyzed for morphological characteristics at the Plant Microstructure Laboratory, Department of Crop Science, Faculty of Agriculture, Universiti Putra Malaysia. The plant’s reproductive organs were viewed using a Hirox light microscope.
Data collection
Morphological traits were measured according to protocol for distinctness, uniformity and stability test of Solanum melongena L. (Solanaceae) (Community Plant Variety Office, 2008). Data collected were including plant height (m), leaf blade length (cm), leaf blade width (cm), petiole length (cm), number of flower, flower diameter (mm), pedicel length (mm), style length (mm), number of fruit, fruit diameter (cm), fruit weight (g), calyx length (cm), calyx width (cm) and number of seed.
Statistical analyses
Analysis of variance (ANOVA) was conducted using the General Linear Model (PROC GLM) of the Statistical Analysis System computer software (SAS)13 version 9.2 (SAS Institute Inc. 2005) to determine the significance of variation among populations. Subsequently, the Least Significant Difference (LSD) was applied for mean comparison of all morphological characteristics. Simple phenotypic correlations among the traits were determined based on the formula suggested by Gomez & Gomez14 and calculated using PROC CORR procedure of SAS. Dissimilarity matrix among the 19 populations was obtained by using Average Euclidean dissimilarity coefficient. Clustering analysis was performed using Unweighted Pair Group Method with Arithmetic Mean (UPGMA) in NTSYS-PC version 2.1software Rohlf.15 Principal Component Analysis (PCA) was performed on the inter-characteristic correlation matrix derived from the standardized morphological characteristics using. The goodness of fit between dissimilarity matrix and dendro gram was estimated using cophenetic correlation coefficient (r) Sokal & Rohlf.16
Germplasm collection
Hundred and thirty individual plants samples of Physalis minima were collected from 11 states of peninsular Malaysia from April 2011 to December 2012 (Table 1) (Figure 1). The sites of collections included farm, plantation area (especially of oil palm (Elaeis guineensis)) and the road site bush. The exact sampling sites with the local site names and code are as indicated in Table 1. Germplasm collection and sampling were done according to method of Hawkes12 where sampling was made every 30 km following state roads in different directions. From each site five to ten plants at the stage of flowering and fruiting were taken. At least 10 samples were collected from every state.
No |
Code |
State |
District |
Location |
Coordinate |
Plant |
1 |
D - 01 |
Kelantan |
Pasir Mas |
Kg. Kubang Batu |
102.169°E, 05.966°N |
5 |
2 |
D - 02 |
Kelantan |
Kota Bharu |
Kg. Kota Jelasin |
102.226°E, 06.087°N |
5 |
3 |
T - 01 |
Terengganu |
Permaisuri |
Kg. Panchor Merah |
102.690°E, 05.565°N |
10 |
4 |
J - 01 |
Johor |
Muar |
Kg. Parit Beting |
102.548°E, 02.069°N |
10 |
5 |
M - 01 |
Melaka |
Melaka Tengah |
Kg. Kandang |
102.303°E, 02.193°N |
10 |
6 |
C - 01 |
Pahang |
Temerloh |
Kg. Bangau |
102.422°E, 03.468°N |
5 |
7 |
C - 02 |
Pahang |
Chenor |
Kg. Batu Gajah |
102.628°E, 03.511°N |
5 |
8 |
N - 01 |
Negeri Sembilan |
Jelebu |
Kg. Kuala Kelawang |
101.968°E, 02.946°N |
5 |
9 |
N - 02 |
Negeri Sembilan |
Seremban |
Kg. Bukit Merbah |
101.947°E, 02.738°N |
5 |
10 |
R - 01 |
Perlis |
Kangar |
Kg. Mata Ayer |
100.254°E, 06.474°N |
5 |
11 |
R - 02 |
Perlis |
Kangar |
Kg. Kubang Perun |
100.256°E, 06.478°N |
5 |
12 |
K - 01 |
Kedah |
Ayer Hitam |
Kg. Matang Paku |
100.264°E, 06.258°N |
5 |
13 |
K - 02 |
Kedah |
Kuala Kedah |
Kg. Masjid Lama |
100.329°E, 06.102°N |
5 |
14 |
B - 01 |
Selangor |
Kuala Selangor |
Pasir Penambang |
101.251°E, 03.348°N |
10 |
15 |
B - 02 |
Selangor |
Serdang |
Farm 2 UPM |
101.704°E, 03.009°N |
10 |
16 |
B - 03 |
Selangor |
Serdang |
Farm 10 UPM |
101.701°E, 03.008°N |
10 |
17 |
A - 01 |
Perak |
Tanjung Malim |
Kg. Ketoyong |
101.510°E, 03.695°N |
5 |
18 |
A - 02 |
Perak |
Slim River |
Kg. Manggis |
101.399°E, 03.825°N |
5 |
19 |
P - 01 |
Pulau Pinang |
Batu Maung |
Kg. Binjai |
100.256°E, 05.284°N |
10 |
Table 1 Geographical distribution of 19 populations of Physalis minima collected from Peninsular Malaysia
Morphological study
One hundred and thirty fresh samples from 19 populations were analyzed for morphological characteristics at the Plant Microstructure Laboratory, Department of Crop Science, Faculty of Agriculture, Universiti Putra Malaysia. The plant’s reproductive organs were viewed using a Hirox light microscope.
Data collection
Morphological traits were measured according to protocol for distinctness, uniformity and stability test of Solanum melongena L. (Solanaceae) (Community Plant Variety Office, 2008). Data collected were including plant height (m), leaf blade length (cm), leaf blade width (cm), petiole length (cm), number of flower, flower diameter (mm), pedicel length (mm), style length (mm), number of fruit, fruit diameter (cm), fruit weight (g), calyx length (cm), calyx width (cm) and number of seed.
Statistical analyses
Analysis of variance (ANOVA) was conducted using the General Linear Model (PROC GLM) of the Statistical Analysis System computer software (SAS)13 version 9.2 (SAS Institute Inc. 2005) to determine the significance of variation among populations. Subsequently, the Least Significant Difference (LSD) was applied for mean comparison of all morphological characteristics. Simple phenotypic correlations among the traits were determined based on the formula suggested by Gomez & Gomez14 and calculated using PROC CORR procedure of SAS. Dissimilarity matrix among the 19 populations was obtained by using Average Euclidean dissimilarity coefficient. Clustering analysis was performed using Unweighted Pair Group Method with Arithmetic Mean (UPGMA) in NTSYS-PC version 2.1software Rohlf.15 Principal Component Analysis (PCA) was performed on the inter-characteristic correlation matrix derived from the standardized morphological characteristics using. The goodness of fit between dissimilarity matrix and dendro gram was estimated using cophenetic correlation coefficient (r) Sokal & Rohlf.16
The result of analysis of variance (ANOVA) revealed significant differences between the 11 states for all quantitative characteristics measured. Table 2 shows results of mean comparisons among 19 P. minima populations collected in Malaysia regardless of their states. Similarly to ANOVA findings, mean comparison showed no significant variation among population within states for all morphological characteristics (Table 2). The range for plant height was from 1.4 to 1.6 cm, with a mean of 1.5 cm. Leaf blade length ranged from 8.5 to 9.7 cm, with a mean of 9.2 cm, while leaf blade width ranged from 6.9 to 8.7 cm, with a mean of 8.0 cm. The range for petiole length was 3.7 to 4.5 cm, with a mean of 4.0 cm. Number of flower ranged from 50 to 63, with a mean of 57.9, while the flower diameter ranged from 1.3 to 1.4 cm, with a mean of 1.3 cm. The range for stamen length was 5.6 to 6.0 mm, with a mean of 6.0, while style length ranged from 8.7 to 9.0 mm, with a mean of 9.0 mm. The number of fruit ranged from 48 to 66, with a mean of 57.5. The fruit diameter was ranged from 1.3 to 1.5 cm, with a mean of 1.4 cm, while the fruit weight ranged from 1.1 to 1.3 g, with a mean of 1.2 g. The calyx length ranged from 3.2 to 3.6 cm, with a mean of 3.5 cm, while the calyx width ranged from 1.5 to 1.9 cm, with a mean of 1.8 cm. The number of seed ranged from 99 to 112, with a mean of 104.5.
|
X1 |
X2 |
X3 |
X4 |
X5 |
X6 |
X7 |
X8 |
X9 |
X10 |
X11 |
X12 |
X13 |
X14 |
X15 |
X16 |
(m) |
(cm) |
(cm) |
(cm) |
(cm) |
(cm) |
(mm) |
(mm) |
(cm) |
(g) |
(cm) |
(cm) |
(cm) |
||||
B - 01 |
1.6a |
10.1a |
8.9a |
5.1a |
64.3a |
1.3a |
1.4a |
7.0a |
9.0a |
73.8a |
1.7a |
1.4a |
2.2a |
4.1a |
2.0a |
115.6a |
B - 02 |
1.6a |
10.1a |
9.0a |
5.1a |
64.7a |
1.3a |
1.4a |
7.0a |
9.0a |
75.0a |
1.6a |
1.4a |
2.2a |
4.1a |
2.1a |
116.1a |
B - 03 |
1.6a |
10.2a |
9.0a |
5.1a |
64.7a |
1.3a |
1.4a |
7.0a |
9.0a |
76.5a |
1.7a |
1.4a |
2.2a |
4.1a |
2.1a |
117.9a |
J - 01 |
1.5b |
9.2c |
8.0d |
4.0c |
57.9b |
1.2b |
1.3c |
6.0bc |
9.0a |
57.5bc |
1.4b |
1.2bcd |
2.2b |
3.5b |
1.8b |
104.5b |
K - 01 |
1.5b |
9.5b |
7.8d |
3.9d |
56.6b |
1.2b |
1.3c |
6.0bc |
9.0a |
53.2d |
1.4c |
1.2cd |
2.2b |
3.4bc |
1.7c |
102.4b |
K - 02 |
1.5b |
9.3b |
8.0d |
3.9d |
59.2b |
1.2b |
1.3c |
5.9bc |
8.9a |
55.2d |
1.3c |
1.1cd |
2.2b |
3.3bc |
1.6c |
102.2b |
D - 01 |
1.5b |
9.5b |
7.8d |
3.9d |
56.6b |
1.2b |
1.3c |
6.0bc |
9.0a |
52.2d |
1.4c |
1.2d |
2.2b |
3.4bc |
1.8b |
102.4b |
D - 02 |
1.5b |
9.3b |
8.0d |
3.9d |
59.2b |
1.2b |
1.3c |
5.9bc |
8.9a |
54.6d |
1.3c |
1.1d |
2.2b |
3.3bc |
1.7b |
102.2b |
M - 01 |
1.5b |
9.1c |
8.3d |
4.1bc |
59.7b |
1.2b |
1.3bc |
5.9c |
9.0a |
58.4bc |
1.4b |
1.2bc |
2.2b |
3.4bc |
1.8b |
107.1b |
N - 01 |
1.5b |
9.0c |
8.7b |
4.2b |
59.0b |
1.2b |
1.3b |
6.0b |
9.0a |
59.0b |
1.4b |
1.2b |
2.2b |
3.3bc |
1.8b |
106.0b |
N - 02 |
1.5b |
9.0c |
8.7b |
4.1b |
59.6b |
1.2b |
1.3b |
6.0b |
9.0a |
59.8b |
1.4b |
1.2b |
2.2b |
3.5bc |
1.8b |
106.0b |
C - 01 |
1.5b |
9.0c |
8.6b |
4.3bc |
58.2b |
1.2b |
1.3b |
6.0b |
9.0a |
59.0bc |
1.4b |
1.2bc |
2.2b |
3.3c |
1.8b |
105.2b |
C - 02 |
1.5b |
9.1c |
8.6b |
4.0bc |
62.6b |
1.2b |
1.3b |
6.0b |
9.0a |
59.2bc |
1.4b |
1.2bc |
2.2b |
3.4c |
1.8b |
105.0b |
P - 01 |
1.5b |
9.0c |
7.8d |
4.2b |
59.3b |
1.2b |
1.3b |
6.0b |
9.0a |
56.1cd |
1.4b |
1.1e |
2.2b |
2.3e |
1.5d |
93.9c |
A - 01 |
1.5b |
9.2c |
7.9d |
4.2c |
56.6b |
1.2b |
1.3c |
6.0bc |
9.0a |
58.8bc |
1.4b |
1.2b |
2.2b |
3.3c |
1.6cd |
105.2b |
S. D |
0 |
0.4 |
0.5 |
0.2 |
4.7 |
0 |
0 |
0.1 |
0.1 |
5.3 |
0 |
0.1 |
0 |
0.1 |
0.1 |
5.7 |
C.V |
4.19 |
2.55 |
3.17 |
3.48 |
7.81 |
1.63 |
0 |
1.29 |
0.65 |
5.45 |
4.18 |
4 |
0 |
4.33 |
7.08 |
4.84 |
Table 2 Mean values and standard deviation for 16 quantitative characteristics measured on 19 Physalis minima populations
Means followed by the same letter in the same column are not significantly different at p ≤ 0.05 based on DNMRT.
X1, Plant height; X2, Leaf blade length; X3, Leaf blade width; X4, Petiole length; X5, Number of flower; X6, Flower diameter; X7, Pedicel length;X8, Stamen length; X9 , Style length; X10, Number of fruit; X11, Fruit diameter; X12, Fruit weight; X13, Stalk length; X14, Calyx length; X15, Calyx width; X16, Number of seed
Results showed low coefficient of variation for plant height, leaf blade length, leaf blade width, petiole length, number of flower, flower diameter, stamen length, style length, number of fruit, fruit diameter, fruit weight, calyx length, calyx width and number of seed with values of 4.2, 2.5, 3.2, 3.5, 4.8, 1.6, 1.3, 0.7, 5.5, 4.2, 4.0, 4.3, 7.1 and 4.8% respectively. This indicates that the samples had values closer to the population mean for each characteristic. There were no C.V. values (NA) for two characteristics which were pedicel length and stalk length. This is due to the homogeneity of the samples. The individuals were similar and no variations detected for that particular traits. The small range of variations observed for the 16 morphological characteristics, indicates that the populations studied had low level of diversity. However, the level of diversity needs to be evaluated at the DNA level since morphological characteristics of populations can be highly influenced by environmental factors Hoey et al.17 Results on phenotypic correlations among characters measured on the populations are presented in Table 3. Based on phenotypic correlation estimates, fruit weight was found to be highly positively correlated (at p≤ 0.01) with stamen length, number of fruit, fruit diameter, petiole length, flower diameter, plant height, leaf blade length, leaf blade width, flower diameter and style length with correlation coefficients of 0.87, 0.86, 0.84, 0.78, 0.72, 0.714, 0.69 and 0.43 respectively. Therefore, for selection purposes to improve populations yield, it is suggested that emphasis should be given on these traits.
|
X1 |
X2 |
X3 |
X4 |
X5 |
X6 |
X8 |
X9 |
X10 |
X11 |
X12 |
X14 |
X15 |
X2 |
0.77** |
||||||||||||
X3 |
0.64** |
0.84** |
|||||||||||
X4 |
0.71** |
0.81** |
0.77** |
||||||||||
X5 |
0.75** |
0.61** |
0.62** |
0.65** |
|||||||||
X6 |
0.65** |
0.75** |
0.66** |
0.88** |
0.58** |
||||||||
X8 |
0.74** |
0.87** |
0.77** |
0.95** |
0.74** |
0.89** |
|||||||
X9 |
0.64** |
0.85** |
0.84** |
0.58** |
0.45** |
0.53** |
0.62** |
||||||
X10 |
0.73** |
0.74** |
0.69** |
0.91** |
0.75** |
0.86** |
0.92** |
0.49** |
|||||
X11 |
0.69** |
0.63** |
0.60** |
0.84** |
0.67** |
0.79** |
0.84** |
0.36** |
0.86** |
||||
X12 |
0.72** |
0.71** |
0.69** |
0.84** |
0.69** |
0.78** |
0.87** |
0.43** |
0.86** |
0.84** |
|||
X14 |
0.59** |
0.72** |
0.68** |
0.71** |
0.53** |
0.66** |
0.78** |
0.46** |
0.75** |
0.66** |
0.83** |
||
X15 |
0.60** |
0.69** |
0.73** |
0.75** |
0.56** |
0.71** |
0.76** |
0.53** |
0.74** |
0.68** |
0.77** |
0.80** |
|
X16 |
0.62** |
0.75** |
0.74** |
0.77** |
0.62** |
0.66** |
0.80** |
0.59** |
0.78** |
0.64** |
0.79** |
0.77** |
0.70** |
Table 3 Simple phenotypic correlation coefficients among characters measured on 19 Physalis minima populations
** = significant at p ≤ 0.01
X1, Plant height; X2, Leaf blade length; X3, Leaf blade width; X4, Petiole length; X5, Number of flower; X6, Flower diameter; X8, Stamen length; X9, Style length; X10, Number of fruit; X11, Fruit diameter; X12, Fruit weight; X14, Calyx length; X15, Calyx width; X16, Number of seed
Relationship among populations
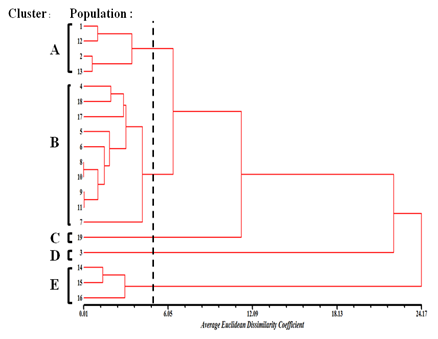
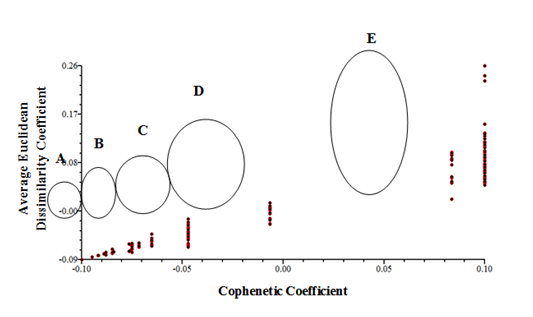
Cluster analysis based on quantitative characteristics using Average Euclidean Dissimilarity correlation coefficient revealed five distinct clusters, designated as A to E (Figure 2). Cluster A consists of Populations 1 and 2 collected from Kelantan and 12 and 13 from Kedah. Cluster B consists of populations 9 populations collected from Johor, Melaka, Negeri Sembilan, Pahang, Perak and Perlis. Meanwhile, cluster C consist of population collected from Penang. The mean values of the populations grouped in cluster A, B and C were found to be moderate for most of the quantitative characteristics measured including plant height, leaf blade length, petiole length, number of flower, number of fruit and number of seed. Cluster D consist of population collected from Terengganu. This cluster had the lowest mean values for all morphological characteristics measured except flower diameter and pedicel length. Populations 14, 15 and 16 (cluster E) collected from Selangor state were found to have the highest mean values for most of the characteristics (Table 4). The plants height was up to 1.6 m, with high number of flowers and fruits. Result showed that populations collected were found to be grouped in three major groups with high (cluster E), medium (cluster A, B and C) and low (D) morphological characteristics. This indicates that, according to the morphological characteristics, three different morphological ranged of P. minima can be found in the 19 populations studied. The cophenetic correlation test was carried out to identify the goodness of fit of cluster analysis to the matrix. The correlation coefficient (r) between the two matrices was 0.93 which was significant based on Mantel test (t value = 5.51, P = 1.00). The result indicated that the dendrogram (Figure 2) represent the original data very well. All populations of Physalis minima were grouped into five different groups according to the morphological characteristics (Figure 3).
Code |
Characteristics |
Mean value of populations within cluster |
||||
Cluster A |
Cluster B |
Cluster C |
Cluster D |
Cluster E |
||
X 1 |
1-Plant height (m) |
1.5 |
1.5 |
1.5 |
1.3 |
1.6 |
X 2 |
2-Leaf blade length (cm) |
9.4 |
9.1 |
9 |
6.5 |
10.1 |
X 3 |
3-Leaf blade width (cm) |
7.9 |
8.4 |
7.8 |
5.3 |
9 |
X 4 |
4-Petiole length (cm) |
3.9 |
4.1 |
4.2 |
3.1 |
5.1 |
X 5 |
5-Number of flower |
57.9 |
59.1 |
59.3 |
54.4 |
64.6 |
X 6 |
6-Flower diameter (cm) |
1.3 |
1.3 |
1.3 |
1.3 |
1.4 |
X 7 |
7-Pedicel length (cm) |
1.2 |
1.2 |
1.2 |
1.2 |
1.3 |
X 8 |
8-Stamen length (mm) |
6 |
6 |
6 |
5 |
7 |
X 9 |
9-Style length (mm) |
9 |
9 |
9 |
8 |
9 |
X 10 |
10-Number of fruit |
53.8 |
58.6 |
56.1 |
45.7 |
75.1 |
X 11 |
11-Fruit diameter (cm) |
1.3 |
1.4 |
1.4 |
1.3 |
1.7 |
X 12 |
12-Fruit weight (g) |
1.2 |
1.2 |
1.2 |
1.1 |
1.4 |
X 13 |
13-Stalk length (cm) |
2.2 |
2.2 |
2.2 |
2 |
2.2 |
X 14 |
14-Calyx length (cm) |
3.4 |
3.4 |
2.3 |
2.5 |
4.1 |
X 15 |
15-Calyx width (cm) |
1.7 |
1.8 |
1.5 |
1.3 |
2.1 |
X 16 |
16- Number of seed |
102.3 |
105.6 |
105.1 |
86.2 |
116.5 |
Table 4 Means of populations within clusters for morphological characteristics
Diversity based on principal component analysis (PCA)
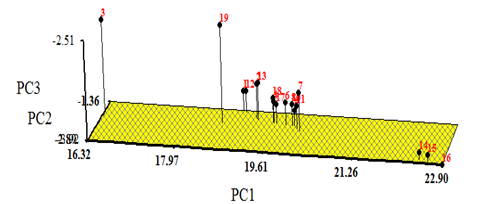
Principal component analysis (PCA) was performed on the morphological data. The given values obtained for the first three principal component scores indicate that they could provide a good description of the data, cumulatively accounting for 94% of the standardized variance (Table 5). The analysis of eigenvectors provides information on the nature of the characteristics responsible for the significant cumulative variation over the first three principal components. The PC1 data set accounted for 81% of the total variation, where all morphological characteristics were associated positively with PC1. Thus, this principal component was responsible for the separation of the populations 14, 15 and 16 from the other populations of P. minima (Figure 4) PC2 accounted for 9% of the total morphological variability and was positively attributable to differences in the plant height (X1), leaf blade length (X2), leaf blade width (X3), style length (X9), calyx width (X15) and number of seed (X16). The floral and fruit characteristics of number of flower (X5), flower diameter (X6), stamen length, number of fruit (X10), Fruit diameter (X11), fruit weight (X12) and calyx length (X14) were found negatively associated with PC2 (Table 3). Thus, this principal component was able to distinguish between populations A, B and C. (Figure 2) PC3 accounted for 4% of the total variation and was positively correlated with the five morphological characteristics, leaf blade width (X3), fruit weight (X12), calyx length (X14), calyx width (X15) and number of seed (X16) but negatively correlated with other 9 characteristics. Thus, PC3 was able to distinguish cluster D from other clusters (Figure 2). (The three axes represent the first three principle components that accounted for 94% of the total variation. Numbers on the graph correspond to the 19 populations of P. minima. Populations 14, 15 and 16 are specified using*).
Axis |
Eigenvalue |
Difference |
Proportion |
Cumulative variance |
PC 1 |
11.3485 |
10.1265 |
0.8106 |
0.8106 |
PC 2 |
1.222 |
0.6551 |
0.0873 |
0.8979 |
PC 3 |
0.5669 |
0.2474 |
0.0405 |
0.9384 |
PC 4 |
0.3194 |
0.0805 |
0.0228 |
0.9612 |
Table 5 Principle component analysis of 16 characteristics associated with 19 P. minima populations
High significant variations among P. minima populations from different states for all morphological characteristics could be due to different environmental factors. Cluster analysis and PCA showed five distinct groups among the 19 populations of P. minima among which Populations 14, 15 and 16 (from Selangor) possessed the highest fresh weight, number of fruits, number of flower and plant height. The populations from same state possessed similar morphological characteristics, indicating the role of environmental factors in morphological variations in P. minima. Level of diversity was high among populations from different states but it was low among populations within each state. This indicates that hybridization of populations from different states can lead to achieve high heterosis for important morphological traits in P. minima. In addition, significant differences in performance indicate that the populations varied substantially among themselves for the traits measured in many aspects, and these differences could be exploited for specific purposes in breeding programs. Since phenotypic values obtained from the populations were influenced by environmental factors, analysis of molecular variation can be performed on the populations as an additional tool to help in the selection process for development of improved populations of P. minlima in Malaysia.
This research was supported by Ministry of Higher Education (MOHE) under FRGS grant (8036). We thank to the staffs from Department of Crop Science, Faculty of Agriculture, UPM who provided insight and expertise that greatly assisted the research.
None.

©2018 Usaizan, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.