Advances in
eISSN: 2573-2862


Research Article Volume 2 Issue 3
Department of Pathology, Universiti Putra Malaysia, Malaysia
Correspondence: Heng Fong Seow, Department of Pathology, Universiti Putra Malaysia, Serdang, Selangor 43400, Malaysia, Tel 60389472387
Received: March 20, 2017 | Published: April 24, 2017
Citation: Khoo SLJ, Yip WK, Seow H. Differential effects of dishevelled 2 and 3 on TCF/LEF- mediated transcriptional activation in wnt-3a-treated breast cancer cell line MDA-MB-231. Adv Cytol Pathol. 2017;2(3):72-79. DOI: 10.15406/acp.2017.02.00021
Background: Dishevelled (Dvl) proteins are central mediators of the Wnt signalling pathway. Previous studies have shown that Dvl1, 2 and 3 can differentially mediate the Wnt signalling pathway and act at different points in the signalling cascade depending on the cell lines used. It is unclear whether these differences also occur in MDA-MB-231 which is a highly invasive fibroblastic triple negative human breast cancer cell line. It is important to determine the differential roles of dishevelled proteins as they could serve as potential useful therapeutic targets in cancer therapy.
Methods: Small interfering RNAs (siRNAs) targeting Dvl1, Dvl2 and Dvl3 in MDA-MB-231 were used to silence the expression of Dvl1, 2 and 3. Western Blot and RT-qPCR were performed to show the silencing of Dvl2 and Dvl3. The invasive and migratory properties of Dvl2 and Dvl 3 were studied using invasion assay and wound healing assay, respectively. This is followed by TCF/LEF-sensitive luciferase reporter assay to study the involvement of Dvl isoforms in TCF-LEF transcriptional activation in Wnt3-stimulated MDA-MB-231. Finally, Western Blot was performed to detect the expression of phosphorylated ß-catenin and total ß-catenin in Dvl2- and Dvl3-silenced cells.
Results: Reduced migration abilities were seen in Dvl2- silenced cells and a 10% and 12% reduction in invasiveness of MDA-MB-231 transfected with siRNAs against Dvl2 and Dvl3, respectively. There was a decrease in Wnt3a-mediated TCF/LEF transcriptional activation with Dvl3 siRNA but an increase with Dvl2 siRNA. Reduction in phospho-ß-catenin levels was seen in Dvl2 but not Dvl3 knockdown. The level of total ß-catenin was sustained in both cases.
Conclusion: In summary, there is involvement of cell migration on Dvl 2. Human Dvl3 plays a significant role in mediating Wnt3a stimulated TCF/LEF transcriptional activation downstream of canonical Wnt signaling pathway. The level of phosphorylated ß-catenin was not changed in Dvl3 silenced cells. MDA-MB231 can be a cell line used to elucidate the differential roles of Dvl 2 and 3. It is important to determine the roles of individual dishevelled proteins and their interacting proteins as they could be potential targets for therapeutic intervention.
Keywords: dishevelled, ß-catenin, mda-mb-231
Dvl, dishevelled; siRNA, small interfering RNA; TCF/LEF, T cell factor/lymphoid enhancer factor; PR, progesterone receptor; ER, estrogen receptor; HER2, human epidermal growth factor receptor-2; APC, adenomatous polyposis coli; CK1, casein kinase; GSK-3ß, glycogen kinase 3 beta; PAGE, polyacrylamide gel electrophoresis; PVDF, polyvinylidene fluoride; HRP, horse-radish peroxidase; DIR, dishevelled interaction region
Breast cancer is the most common cancer amongst women which can result in death. In 2012, more than 400,000 women in Europe died from breast cancer.1 A total of 32,340 new cases of invasive breast cancer and 39,620 of breast cancer deaths have been reported in the United States in 2013.2 The Wnt signaling pathway plays an important role in cancer metastasis.3–5 A previous study has shown an increased frequency of nucleocytoplasmic β-catenin expression in 56% of ductal carcinoma in-situ, indicating activation of canonical Wnt signaling in the early stages of human breast neoplasia.6 Basal-like breast cancer, also known as triple negative breast cancer, is negative in expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor-2 (HER2) is difficult to treat.7 Basal-like breast cancer has been found to be positive for cytoplasmic and nuclear accumulation of β-catenin by immunohistochemical staining.8 Nuclear and cytoplasmic accumulation of β-catenin in basal- like breast cancers is predictive of worse overall survival.4,8 Inhibition of the Wnt pathway reduced stem-like cell activity in patient- derived metastatic breast cancer cells.9 Thus, these studies provide evidence to support the potential of Wnt-targeted therapeutics for breast cancer patients.4,9
Dishevelled (Dvl) proteins are central mediators of the Wnt signalling pathway which plays an important role in embryonic development, generation of cell polarity and cell specification.10 Aberrant activation of the Wnt signal transduction pathway has been shown to lead to tumorigenesis.11 Wnt proteins initiate three distinct pathways, namely, canonical, non-canonical or planar cell polarity and Wnt-Ca2+ pathway. Dishevelled proteins are components of all three signalling pathways. In the absence of Wnt, a multiprotein destruction complex consisting of β-catenin, Axin, adenomatous polyposis coli (APC), casein kinase (CK1) and glycogen kinase 3 beta (GSK-3β) is formed. This complex facilitates the phosphorylation and ubiquitination of β-catenin and results in proteosomal degradation of β-catenin12 and thus, the level of cytoplasmic β-catenin is low. Binding of Wnt ligands to Frizzed and low density lipoprotein-related protein 5 and 6 results in recruitment of dishevelled which binds to axin.13 Dvl promotes the stabilization of β-catenin by binding with Axin at the cell membrane. This disrupts the formation of the β-catenin destructive complex allowing the β-catenin to accumulate and translocate to the nucleus.14 In the nucleus, β-catenin binds to the transcription factor TCF/LEF (T cell factor/lymphoid enhancer factor) and activates the expression of Wnt target genes such as c-myc, c-jun and cyclin D1.15 More recently, it has been shown that two Forkhead box transcription factors, FOXK1 and FOXK2 activate Wnt/β-catenin signalling by translocating dishevelled into the nucleus.16 There are three members of the dishevelled proteins, namely, Dvl1, 2 and 3 and all three dishevelled proteins were shown to interact with FOXK proteins in HEK293T (human embryonic kidney cell with SV40 T-antigen) and HT29 (colon cancer) cells. Thus, dishevelled not only acts as the scaffold protein between Frizzled receptors and the β-catenin destruction complex but also plays a pivotal role in the nucleus by mediating the formation of the β-catenin/TCF/LEF transcriptional complex.
Dishevelled are over expressed in various cancers including lung,17,18 prostate,19 breast,20,21 cervical cancers,22 and gliomas.23 Studies with SW480, a colon cancer cell line showed that siRNA against Dvl1/3 and Dvl2 inhibited TCF/LEF transcriptional activation and there was marked accumulation of Dvl3 in the nuclei of colon cancer tissues.24 In mouse F9 cells, knockdown of Dvl1 and Dvl3 showed a reduction of Wnt3a stimulated TCF/LEF transcriptional activation but least effect was observed with knockdown of Dvl2.25 Over expression of Dvl1 but not Dvl3 significantly enhanced TCF/LEF transcriptional activity in lung cancer cell lines, A549 and LTEP-α-2.17 Zhao et al.17 also showed that Dvl1 enhanced cell invasiveness via the canonical Wnt signalling pathway in contrast to Dvl3 which acted via the non-canonical Wnt signalling pathway. Thus, three dishevelled proteins can differentially mediate the Wnt signalling pathway and act at different points in the signalling cascade depending on the cell lines used. It is unclear whether these differences also occur in MDA-MB-231 which is a highly invasive fibroblastic triple negative human breast cancer cell line. In this study, recombinant human Wnt3a was used to stimulate TCF/LEF transcriptional activation in MDA-MB-231 cell line. Small interfering RNAs (siRNAs) targeting Dvl2 and 3 isoforms were used to determine the role of Dvl2 and 3 isoforms in mediating TCF/LEF transcriptional activation in the canonical Wnt signaling pathway.
Cell culture
MDA-MB-231 human breast cancer cell line was used in this study. Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, USA) was used supplemented with 10% fetal bovine serum (FBS), 2g/L sodium bicarbonate, penicillin (100units/ml), and streptomycin (100µg/ml) (GIBCO®, Invitrogen Corp, Carlsbad, CA) and kept in a humidified incubator under the temperature of 37oC with 5% of carbon dioxide (CO2).
Transfection of siRNA
Small interfering RNA (siRNA) used to target Dvl1, Dvl2 and Dvl3 mRNAs were purchased from Qiagen (Qiagen Group, MA, USA). Hs_DVL1_6 (Cat no. SI02633183) (CTGGAGTAGGGATCTAATTTA), labelled as “D1_6”, Hs_DVL2_4 (Cat no. SI00063455) (CTGGGAGATTTGGGCCATGTA), labelled as “D2_4” and Hs_DVL3_7 (SI03095463) (CTGCGGGAGATTGTGCACAAA), labelled as “D3_7”, were used to silence Human Dvl2 and Human Dvl3 respectively. Allstars Negative Control siRNA (Cat no. 1027280), containing a scrambled siRNA sequence, acted as a negative control. MDA-MB-231 cell lines were transfected with Lipofectamine 2000 (Cat no. 1166087) (Life Technologies Inc., Rockville, MD, USA).
Reverse-transcription quantitative real time PCR
Human breast cancer cell line MDA-MB-231 was transfected with Dvl siRNAs. Samples were allowed to incubate for 48 hours after transfection. Forty eight hours after transfection, RNA from respective samples were extracted using Genejet RNA Purification Kit (Cat no. K0731) (Thermo Fisher Scientific, Vilnius, Lithuania) and steps involved in RNA extraction were done according to manufacturer’s instructions. Following RNA extraction, DNase treatment was performed on RNA samples to digest carryover genomic DNA content after RNA purification. DNAse1, RNase free kit (Cat no. EN0521) (Thermo Fisher Scientific, Vilnius, Lithuania) was used to perform genomic DNA digestion and steps involved in DNase treatment were done according instructions from manufacturer.
Complementary DNA (cDNA) that was produced from reverse transcription was used as a template for Quantitative Real Time PCR. Maxima SYBR Green qPCR Master Mix (2X) was purchased from Thermo Scientific (Cat no. K0251) (Thermo Fisher Scientific, Vilnius, Lithuania) is the premix Master Mix kit which is used for Quantitative Real Time PCR experiments in this experiment. Primers were purchased from AIT Biotech (Singapore) to detect Human Dvl1 mRNA ( DVL1-F 5'-CCCCTCCTTCCACCCAAATG-3', DVL1-R- 5'-GTGACTGACCATGGACTCCG-3'), Human Dvl2 mRNA (DVL2-F2- 5'- TGAGCAACGATGACGCTGTG -3', DVL2-R2- 5'- AACCTGGATAGGCTGGGAAG -3') and Human Dvl3 mRNA (DVL3-F-5'-TGGACGACGATTTCGGAGTG-3',DVL3-R-5'-GCTCCGATGGGTTATCAGCA-3'). Human β-actin primer pairs (ACTB-F- 5’-GACTTAGTTGCGTTACACCCTTTC-3’,ACTB-R-5’-TGCTGTCACCTTCACCGTTC-3’) was used to amplify the presence of human β-actin mRNA levels which function as reference gene. All primers in this study were shown to share the same annealing temperature of 58oC. Differences between siRNA transfected samples and control (scrambled siRNA sequence) were tested by one way analysis of variance (ANOVA) followed by Dunnett multiple comparison test.
Western blot
The following primary antibodies were used: anti human Dvl1 Antibody (AF3316, R&D Systems Europe, Abingdon, UK), anti Human Dvl2 (sc-8026) & anti-human Dvl3 Antibodies (sc-8027) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). anti-phospho-β-catenin antibody (#9651, Cell Signalling Technology Inc, Danvers, MA, USA), anti-total β-catenin (610153, BD Transduction Laboratories, San Jose, CA,USA), and anti-human α-tubulin (Lab Vision Corporation, Fremont, CA, USA).Cells harvested after transfection were briefly rinsed with cold Phosphate Buffered Saline (PBS) before lysis in 1X SDS sample buffer [62.5mM Tris-HCl (pH6.8), 2%SDS, 10%glycerol, 100mM DTT]. After agitation in a shaking incubator, the cell lysates were homogenized by repeated pipetting using a micropipette.
Polyacrylamide Gel Electrophoresis (PAGE) was performed by loading a total 20µg of cell lysate into each well. This process was followed by electro-transfer onto a Polyvinylidene fluoride (PVDF) membrane. The membrane was incubated with blocking buffer [Tris buffered saline containing 5% BSA (bovine serum albumin)] for 1h. After blocking, the membrane was incubated with the desired antibody overnight under 4oC and then Horse-Radish Peroxidase (HRP) conjugated anti-mouse or anti-rabbit secondary antibody for 1h. After incubation, a mixture of chemiluminescent substrate and chemiluminescent enhancer (SuperSignal® West Femto Maximum Sensitivity Substrate; Thermo Fisher Scientific, Rockford, IL) and images of bands were visualized and captured using Fluo Chem TM 5500 imaging system (Alpha Innotech corp, San Leandro, CA). Images of blots that were captured were used for analysis. Densitometric analysis was performed using ImageJ software (National Institutes of Health: Bethesda, MD, USA).
Migration assay (Wound Healing Assay)
Wound healing assay for siRNA transfected cells was performed only on MDA-MB-231 breast cancer cell line. A total of 75000 cells were seeded on each well and cells were transfected with siRNA targeting Human Dvl 2 & 3. Transfected cells were incubated for two days after transfection in order for the cells to achieve confluency. RPMI media of confluent transfected cells, which contains 10% FBS were then removed and wound was introduced on each well using white pipette tips. Detached cells and cell debris were removed by washing with PBS. After washing, each well was replaced with fresh RPMI media with either 10% FBS or 1% FBS depending on the experiment. The same inverted fluorescence microscope was used to capture images as with wound healing assays involving treatment with Wnt inhibitors. Percentage of recovery was calculated by dividing reduced length of gap captured on a particular time point with the length of gap at 0 hours (the beginning time point), multiplied by 100. Experiment was repeated for three times. Differences between siRNA transfected samples and control (scrambled siRNA sequence) were tested by one way analysis of variance (ANOVA) followed by Dunnett multiple comparison test.
Invasion assay
MDA-MB-231 cell line is seeded on 24-well plate with the density of 50,000 cells per well and were transfected with siRNA targeting Human Dvl 2 & 3 respectively. Twenty four hours after siRNA transfection, cells were trypsinized and was resuspended with RPMI containing 2.5% FBS with penicillin (100units/ml), and streptomycin (100µg/ml). Cells that were resuspended were collected on microtubes followed by centrifugation for 5 minutes. Supernatant was removed after centrifugation and the residual pellet was resuspended with RPMI media without FBS (serum free). Fifty thousand cells is taken from respective samples were transferred to the cell insert, placed in a companion of a 24-well plate (FALCON®, BD Biosciences) which had been coated with Matrigel, ultimately forming the upper chamber. On the other hand RPMI media containing10% FBS was added to the lower chamber, which functions as an attractant. Samples were allowed to invade from the upper chamber to the lower chamber for 36hours.
After 36 hours of cell invasion, cell insert was removed and inserts from respective samples were fixed with 95% ethanol for 10minutes under -20oC. Cell insert was then washed with PBS and later RNase treatment was performed on the lower membrane of the insert for ten minutes. Lower surface, which contains the invaded cells, were stained with 1µg/ml PI (Propidium iodide) (Sigma-Aldrich, St. Louis, MO). Membrane was cut with the help of scalpel and it is transferred to a glass slide with the invaded cells facing the base of the glass slide. A drop of fluorescent mounting media (KPL) was applied to the bottom of glass slide. A drop of DAPI (4’,6-diamine-2’-phenylindole; KPL, Gaithersburg, MD), which had been mixed with fluorescent mounting media (KPL), forming the final concentration of 1µg/ml, was applied to the membrane that faces up, containing non-invaded cells. A cover slip was used to cover the membrane and then viewed under the microscope.
Twelve random fields of a single membrane under 200X magnification were chosen in a zig-zag pattern. Images of PI and DAPI stained cells were captured separately on the same field using a CCD camera (ColorView12; Olympus Soft Imaging Solutions GmbH, Munster, Germany), installed on an Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan) attached to a computer. Percentage of cells invaded were calculated by dividing the average number of invaded cells with the average of total cells (non-invaded & invaded cells), and multiplied by 100. Experiments were repeated three times. Differences between siRNA transfected samples and control (scrambled siRNA sequence) were tested by one way analysis of variance (ANOVA) followed by Tukey Kramer multiple comparison test.
TCF/LEF- sensitive luciferase reporter assay
MDA-MB-231 cells were transfected with Dvl siRNAs in 96 well plates. After 48h, they were transfected with reported plasmids M50 Super 8x Topflash or M51 Super 8X Fopflash as negative control (gifts from Dr Randall Moon)18" and pRL-TK Renilla plasmid (Promega Corporation, Madison WI, USA) as endogenous control using Turbofect in-vitro tranfection reagent. (R0531, Thermo scientific, Waltham, MA, USA) The transfected cells were incubated for 24h before stimulation for 6h with Wnt 3a (100ng/ml). The dose used in Wnt3a stimulation in this study was determined from a study performed by Matsuda et al.26 The cells were lysed using 20µl of Passive Lysis Buffer from the Dual Luciferase® Reporter Assay System Kit (Cat no. E1960) (Promega Corporation, Madison WI, USA) 10µl of cell lysates were transferred to the GloMax®-96 micro plate lumino meter (Pro mega Corporation, Madison WI, USA). Fiftyµl of Luciferase Assay Reagent II was added and placed in the luminometer to detect Luciferase enzyme signal. After detection of the Luciferase signals, 50µl of Stop & Glo Reagent, was added to the luminometer plate to measure Renilla Luciferase activity. A normalized value of Luciferase signal is generated by dividing the signal value of Luciferase with the signal of Renilla Luciferase. All experiments were repeated three times in duplicates. Differences between siRNA transfected samples and control (scrambled siRNA sequence) were tested by one way analysis of variance (ANOVA) followed by Dunnett multiple comparison test.
Statistical analysis
Statistical analysis for Luciferase Reporter Assay were performed using GraphPad InStat version 3 for Windows (Graphpad Software, Inc, San Diego, CA). Graphs shown in this study were done using Graphpad Prism 3.01 for Windows (Graphpad Software Inc, San Diego, CA).
Small interfering RNA (siRNA) targeting Dvl1, 2 and 3 knocked down Dvl1, 2 and 3 mRNA and Dvl2 and 3 protein expression
Reverse Transcriptase Quantitative Real Time PCR were performed to ascertain the reduction of mRNA levels of the respective Dvl1, 2 and 3 mRNAs with the presence of a calibrator (si-SCR) in which MDA-MB-231 cell line was transfected with siRNA containing scrambled sequence. There is a significant reduction (p<0.05) in Dvl1, 2 and 3 mRNA levels (Figure 1A, 2A & 2C), indicating a significant knockdown of Dvl1, 2 and 3.
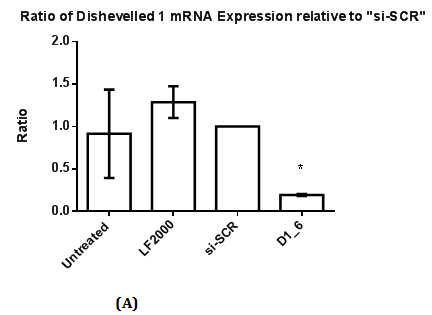
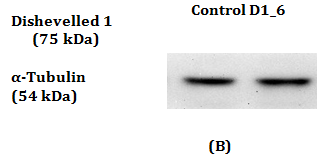

Figure 1 RT-qPCR and Western Blot with antibody against Dvl1 in MDA-MB-231.
1(A): The ratio of Dishevelled 1 mRNA expression relative to control (si-SCR). Statistical analysis was performed using One Way ANOVA followed by Dunnett Multiple Comparison Post hoc test. *p<0.05 vs. “si-SCR” (control).
1(B): Dvl1 protein was absent in Control and also in siRNAs against Dvl1 labelled “D1_6”.
1(C): Expression of Dvl1 protein was detected in other cell lines. Absence of band of Dvl1 protein indicates the absence of Dvl1 isoform on MDA-MB-231 cell lines.
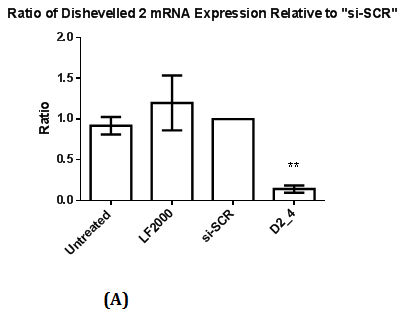
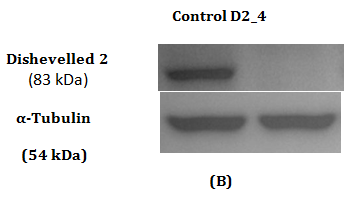
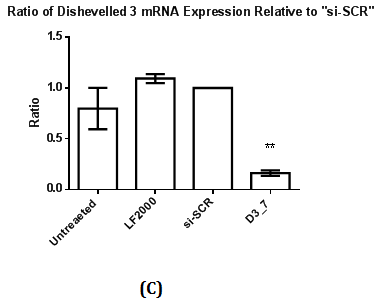

Figure 2 Assessment of Dvl2 and 3 siRNA on Dvl2 and 3 mRNA and protein levels as measured by RT-qPCR and Western Blot.
2(A): The ratio of Dvl2 mRNA expression relative to control (si-SCR) .Statistical analysis was performed using One Way ANOVA followed by Dunnett Multiple Comparison Post hoc test. **p<0.01 vs. “si-SCR” (control).
2(B): Western Blot of Dvl2 protein.
2(C): The ratio of Dvl3 mRNA expression relative to control (si-SCR) .Statistical analysis was performed using One Way ANOVA followed by Dunnett Multiple Comparison Post hoc test. **p<0.01 vs. “si-SCR” (control).
2(D): Western Blot of Dvl3 protein.
Western Blot was conducted to detect the reduction in expression of respective Dvl1, 2 and 3 proteins. A reduction in intensity of Dvl2 and Dvl3 proteins was observed indicating effective gene silencing with siRNA against Dvl2 and 3 (Figure 2B & 2D). Dvl1 protein was not detected on transfected MDA-MB-231 cell lines transfected regardless of cell lines transfected with siRNA against Dvl1 or siRNA harboring scrambled sequence (Figure 1A). To verify that the absence of Dvl1 protein is not due to siRNA transfection Western blot of lysate from untransfected MDA-MB-231 and other cell lines such as K562, T47D and MCF7 was performed The absence of Dvl1 protein in MDA-MB-231 cell line was confirmed (Figure 1C). Due to the absence of Dvl1 in MDA-MB-231, only individual siRNAs targeting Dvl2 (D2_4) and Dvl3 (D3_7) were used in the consecutive experiments.
Effect of Dvl2 and Dvl3 knockdown on migration and invasion
The invasive and migrative effects on Dvl2 and Dvl3 knockdown cells were studied. Invaded MDA-MB-231 cells are located at the lower membrane whereas the non-invaded MDA-MB-231 cell lines are located at the upper membrane. Percentage of invaded cells were obtained by dividing the number of invaded cells against the total number of cells (invaded+non invaded) in the same field. A mean percentage of invaded cells were obtained based on the percentage of invaded cells generated from 20 fields from the same membrane with cells that were treated with the respective Dvl siRNAs. The invasive ability of the MDA-MB-231 cell line after siRNA transfection targeting Dvl2 (D2_4) and Dvl3 (D3_7) was studied and “si-SCR” which contains scrambled siRNA sequence, served as a calibrator. There was a 10% and 12% reduction in invasiveness of MDA-MB-231 transfected with siRNAs against Dvl2 and Dvl3, respectively (Figure 3).
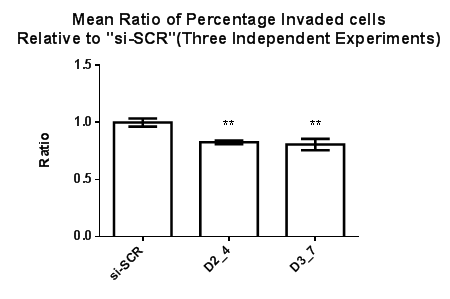
Figure 3 Effect of siRNAs targeting Dvl2 and 3 on MDA-MB-231 cell invasion. The percentage of invaded cells expressed as the ratio of invaded MDA-MB-231 cells transfected with Dvl2 and 3 siRNAs as compared to “si-SCR” (negative control), Statistical analysis was performed using One Way ANOVA followed by Tukey Kramer Multiple Comparison Post hoc test. **p<0.01 vs. “si-SCR” (Control).
Wound healing assay was performed to study the migration ability of cells after siRNA transfection at 48 hours. Transfected cells were allowed to be incubated under normal (10% v/v) and low concentration of FBS (1% v/v). siRNAs targeting Dvl2 (siRNA D2_4) and Dvl3 (D3_7) showed a difference in wound recovery within 48 hours. In cells transfected with Dvl3 siRNA (D3_7), gap of the wounds inflicted were recovered at the final time point (48th hour) of the experiment regardless of high or low concentration of FBS (Figure 4A & 4B). However, for cells transfected with Dvl2 siRNA, wounds only achieve 49% and 74% of recovery towards the final time point under conditions with low (1% v/v) (Figure 4A) and high concentration (10% v/v) (Figure 4B) of FBS, respectively.
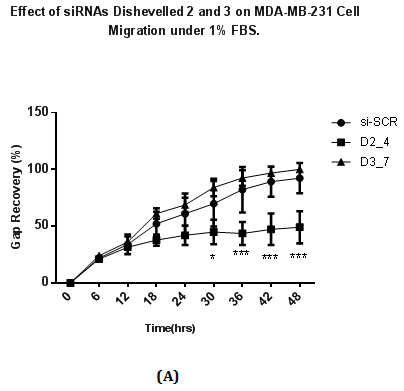
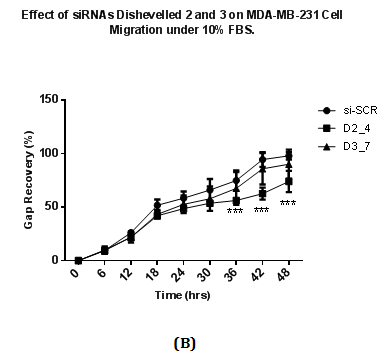
Figure 4 Effect of siRNAs to Dvl 2 and 3 on MDA-MB-231 cell migration with cell culture media containing containing
4(A): 1% v/v FBS
4(B): 10% v/v FBS.
A total of 75000 MDA-MB231 cells were seeded on each well and transfected with siRNAs at a final concentration of 25nM and in RPMI plus 10% FBS. Transfected cells were incubated for two days after transfection and conditioned media is removed. A wound is introduced in each well using white pipette tips. Detached cells and cell debris were removed by washing with PBS. After washing, each well was replaced with fresh RPMI media with either 10% FBS or 1% FBS. The percentage of recovery was calculated by dividing reduced length of gap captured on a particular time point with the length of gap at 0 hours (the beginning time point), multiplied by 100. Results were analysed using Two Way ANOVA followed by Bonferrini post hoc test.
*p<0.05 vs. si-SCR,
***p <0.001 vs. si-SCR.
Silencing of Dvl3 reduces Wnt3a-stimulated TCF/LEF-mediated transcriptional activation
TOP flash and FOP flash Luciferase Assay was performed to measure the transcriptional activation of TCF and LEF upon β-catenin binding to TCF/LEF, leading to up regulation of expression of downstream targets such as Cyclin D1 and c-myc.10 To determine the role of Dvl in TCF/LEF transcriptional activation, knockdown of respective Dvl isoforms was performed (Figure 5). Stimulation with human recombinant Wnt3a protein is required to activate TCF and LEF transcriptional activation in MDA-MB-231 cell line. MDA-MB-231 cell lines transfected with FOP flash plasmid DNA (sample labelled “Untreated FOP”) contains mutated sequences of TCF/LEF binding sites. It acts as a negative control for TCF/LEF transcriptional activation. A five-fold increase in TCF/LEF transcriptional activity in Wnt3a-stimulated cells transfected with siRNA against scrambled sequence (normalized value of 1) was observed as compared to a non-Wnt3a stimulated scrambled siRNA transfected (Figure 5). No increase in Luciferase signal was observed in Wnt-3a stimulated cells transfected with FOP flash plasmid DNA hence showing that the negative control does not affect TCF/LEF mediated transcriptional activation.
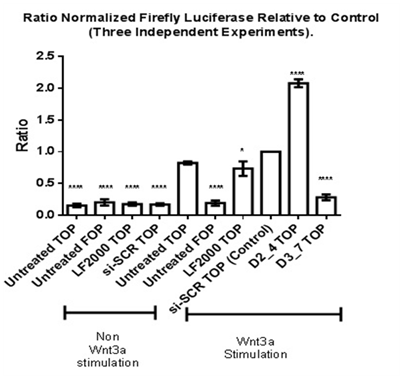
Figure 5 Effect of siRNAs against Dvl1 and Dv3 TCF/LEF transcriptional activation in Wnt3a-stimulated MDA-MDA231. Statistical analysis was performed using One Way ANOVA followed by Dunnett Multiple Comparison Post hoc test.
*p<0.05 vs. Wnt3a Stimulated “si-SCR TOP” (Control).
****p<0.0001 vs. Wnt3a Stimulated “si-SCR TOP” (Control).
Wnt3a-stimulated MDA-MB-231 cell lines which were transfected with Dvl3 siRNA (D3_7) showed a low normalized TCF/LEF Luciferase reporter activity as compared to other samples. This low reporter activity is statistically significant when compared to the calibrator (si-SCR-TOP). In contrast, there was a significant increase in TCF/LEF transcriptional activity in MDA-MB-231 cells transfected with Dvl2 siRNA (siRNA D2_4), in which the effect of Luciferase activity is different compared to the knockdown of Dvl3.
Detection of β-catenin and phospho-β-cateninin MDA-MB-231 cells transfected with siRNA against Dvl3
Western Blot was conducted to observe the changes of β-catenin, which is the key molecule of canonical Wnt signaling pathway. The phosphorylated form of β-catenin indicates a higher tendency of β-catenin degradation, which prevents cytoplasmic accumulation and nuclear translocation of β-catenin for TCF and LEF binding.27 The antibody used in this experiment specifically targets phosphorylated amino acids at Serine 33, 37 and Threonine 41 in β-catenin. These amino acid residues are important for β-catenin phosphorylation followed by ubiquitinylation, which ultimately facilitates β-catenin degradation.28 We hypothesized that knockdown of Dvl isoforms promotes the formation of β-catenin degradation complex and hence result in a higher expression of phosphorylated β-catenin. Based on the results shown on Western Blot (Figure 6), there is decrease in the level of phosphorylated β-catenin in knockdown of Dvl2 but not with knockdown of dvl3. Hence, this indicates that knockdown of Dvl2 can lead to reduced β-catenin phosphorylation and possibly lead to less degradation leading to nuclear translocation of β-catenin and TCF/LEF transcriptional activation. Sustained levels of phospho-β-catenin and total β-catenin in Dvl3 siRNA transfected cells suggest that the reduced TCF/LEF transcriptional activation observed was not due to the loss of β-catenin but due to other mechanisms.
Effect of Dvl2 on cell migration
In the wound healing assay, full wound recovery control (si-SCR) was observed regardless of FBS concentrations (10% v/v versus 1% v/v).This indicates that migration of MDA-MB-231 cells was not dependent on FBS concentrations. Divl2 knockdown MDA-MB-231 cells failed to achieve full wound recovery under both concentrations and were only able to achieve approximately half of full wound recovery towards the endpoint of the experiment suggesting that Dvl2 plays a role in migration of MDA-MB-231. Previous studies had shown that knockdown of Dvl2 in MDA-MB-231 cell line results in significant reduction on Wnt5a-induced wound healing assay and this involves the non-canonical Wnt signaling pathway.21,29 Also, another study done by Yin et al. shows that degradation of Dvl1, 2 and 3 by expressing DACT1 (Dapper) results in reduced migration of MDA-MB-231 cell line.30 A study conducted by Yang et al. on knockdown of Dvl2 using LNCAP prostate cancer cell line showed reduced cell migration. Downstream Western Blot studies showed that knockdown of Dvl2 expression on LNCAP cell lines resulted in reduced expression of Wnt3a, AR (Androgen Receptor), MMP-2 and MMP-9 expression.31 MMPs (Matrix metalloproteinases) play a role in cell migration where activation of MMPS are essential for cells to migrated by rearrangement of extracellular.32 In addition, MMPs are one of the downstream target genes of Wnt β-catenin pathway.33 Further downstream studies required in order to study the mechanisms involved on Dishevelled 2 towards migration of MDA-MB-231 cell lines.
Differential effects of Dvl2 and Dvl3 knockdown in canonical Wnt signaling pathway
The Wnt signaling pathway has been shown to be involved in triple negative breast cancers (TNBC), whereby, aberrant β-catenin expression was found.34 Studies have shown that components of the Wnt signaling pathway such as LRP6 and FZD7, are involved in the upstream part of the canonical Wnt signaling pathway. The canonical Wnt signalling pathway play an important role in cell proliferation and TCF/LEF mediated transcriptional activation on TNBC.35,36 MDA-MB-231 is a highly invasive, “basal type” fibroblastic breast cancer cell line,37 undergoes autocrine Wnt signaling which down regulates E-cadherin expression at cell-cell borders.38
As one of the components involved in canonical Wnt signaling pathway, Dvl was found to be localized in the cytoplasm. Cytoplasmic protein Dvl is recruited to the cell membrane and binds to Axin.13 Such activation of Dvl results in Axin being recruited to the plasma membrane.38 Wnt3a was used to stimulate TCF/LEF-mediated transcriptional activation in MDA-MB-231 cells.39 Wnt3a has been shown to activate canonical Wnt signaling pathway where stimulation with Wnt3a increases β-catenin protein39,40 and also result in translocation of β-catenin to the nucleus.41 Activation of β-catenin due to Wnt3a stimulation on MDA-MB-231 also resulted in increased cell migration based on the study performed by Matsuda et al.26
Previous studies have shown that Dvl1 and 3 isoforms play a role in TCF/LEF transcriptional activation in small cell lung cancer.17 Individual siRNA transfection had been performed on mouse F9 cells under Wnt3a stimulation where individual knockdown of Dvl1, 2 and 3 results in individual reduction of TCF/LEF transcriptional activation.25 Mutation of APC results in aberrant activation of canonical Wnt β-catenin mediated transcriptional activation in diseases such as colorectal cancer. Simultaneous knockdown of Dvl isoforms in APC mutated colorectal cancer cell line SW480 resulted in reduced TCF/LEF mediated transcriptional activation.24 Simultaneous of Dvl2 and Dvl3 suppressed FOXK mediated-canonical Wnt signaling pathway by suppressing FOXK- induced TOP Flash Luciferase activity under stimulation of lithium chloride on HEK293T cells.16
Our results show that knockdown of Dvl3 by siRNA against Dvl3 (D3_7) was effective in suppressing Wnt3a-stimulated TCF/LEF transcriptional activation. Hence, this supports the role of Dvl3 isoform in TCF/LEF transcriptional activation. In contrast, we observed that knockdown of Dvl2 isoform through siRNA D2_4 increased TCF/LEF transcriptional activation suggesting that Dvl2 acts as a negative regulator in Wnt3a stimulated transcriptional activation. A recent study by Jiang et al.42 indicate a new function of Dishevelled on HEK293 cells where Dvl interacts with ZNRF3 and RNF43, resulting in degradation of FZD.42 ZNRF3 and RNF43 play a role as negative feedback regulators induced by Wnt β-catenin signalling in which both ligases promotes ubiqutinylation and lysosomal degradation of Wnt receptors FZD and LRP6.43,44 Deletion studies on Dishevelled Interaction Region (DIR) between ZNRF3 and RNF43 on HEK293 cells shows increased TCF/LEF Luciferase Reporter Assay, hence indicating that Dishevelled can act as a negative regulator in Wnt β-catenin pathway.42
Involvement of Dvl 2 and 3 in inhibition of β-catenin degradation complex
Dvl plays a role in Wnt β-catenin pathway by inhibiting formation of β-catenin degradation complex by interacting with Axin.13 Previous studies have shown that deletion of the DIX domain (one of the three domains on Dvl protein) results in phosphorylation of β-catenin which ultimately results in reduced cytosolic β-catenin.13,37 We hypothesized that knockdown of Dvl3 results in enhanced β-catenin degradation due to increased tendency for formation of β-catenin degradation complex. However, our results show that there is no significant increase in phosphorylated β-catenin levels in cells transfected with siRNA against Dvl3. The level of phosphorylated β-catenin, which is an indication of a higher tendency in β-catenin degradation, remained the same when compared to Wnt3a-stimulated cells transfected with siRNA against scrambled sequence (control). Thus, the same level of expression in phosphorylated form of β-catenin indicates that reduced TCF/LEF transcriptional activity is not due to β-catenin degradation. This indicates that there are other mechanisms which have contributed to the reduced TCF/LEF transcriptional activity.
Western Blotting results shows a reduced level phosphorylated β-catenin in Dvl2-silenced cells. This can lead to a lesser tendency of β-catenin degradation and increased β-catenin accumulation to the nucleus and hence activation of Wnt3a-mediated TCF/LEF transcriptional activation. Thus, this is an additional role of Dvls other than stabilization of β-catenin by binding with Axin at the cell membrane.14 Other than the possibility where Dvl2 might play a role as a negative regulator in Wnt β-catenin pathway (as shown by the luciferase gene reporter assay), it is possible that there are other compensatory mechanisms which inhibit β-catenin degradation or increase accumulation of β-catenin independent of Dvl2. Further investigation is required to study the mechanisms involved in increased TCF/LEF transcriptional activation.
Non-involvement of Dishevelled in the cytoplasm but in the nucleus during activation of Wnt β-catenin pathway was observed on a study performed by Itoh et al.45 where there is increased translocation of Dvl2 isoform at the nucleus on MCF7 breast cancer cell line under Wnt3a stimulation.45 Translocation of Dvl 2 to the nucleus was also observed on mouse F9 cells where the expression of Dvl 2 of the nuclear fraction increases after Wnt3a stimulation based on Western Blot, together with coupled interaction of Axin with Dvl2 and β-catenin during activation of the canonical pathway by Wnt3a in mouse F9 cells.46 Truncated function of APC (Adenomatous Polyposis Coli), which results in increased β-catenin accumulation was seen on cells line such as SW480. A study performed by Gan et al.24 shows that simultaneous knockdown of Dvl 1, 2 3 isoforms through transfection of siRNAs targeting Dvl1, 2 and 3 into SW480 colorectal cancer cell line resulted in reduced TCF/LEF-mediated transcriptional activation but did not result in the loss of β-catenin.24 The same study further shows that Dvl3 was found to interact with c-Jun to bind to c-myc promoter, in which c-myc is one of the downstream target genes for canonical Wnt signaling pathway.24 A recent study conducted by Wang et al.16 shows that Fork head box (FOX) transcription factors FOXK1 and FOXK2 plays a role as Dvl-associated proteins in which both FOX transcription factors active canonical Wnt signaling pathway by promoting DVL nuclear translocation.16 Future studies to determine the differences underlying the role of Dvl2 and 3 in the cytoplasm and in the nucleus including TCF/LEF transcriptional activation in MDA-MB-231 is highly recommended.
In summary, our data shows that there is a differential role of Dvl2 and 3 in MDA-231 cell line. Inhibition of cell migration was accomplished with siRNA against Dvl2 but not Dvl3. Human Dvl3 plays a significant role in mediating Wnt3a-stimulated TCF/LEF transcriptional activation in MDA-MB-231 as inhibition of TCF/LEF transcriptional activation is achieved with siRNA against Dvl 3 but not Dvl2. The level of total ß-catenin was not changed in Dvl3 silenced cells. MDA-231 can be a cell line used to elucidate the differential roles of Dvl 2 and 3. It is important to determine the roles of individual dishevelled proteins and their interacting proteins as they could be potential targets for therapeutic intervention.
This work is supported by a grant from the Malaysian Ministry of Science, Technology and Innovation (project number 02-01-04-SF1312).
The author declares no conflict of interest.

©2017 Khoo, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.