Advances in
eISSN: 2573-2862


Research Article Volume 2 Issue 2
1Division of Cancer and Stem Cell, University of Nottingham, UK
2Department of Morbid Anatomy & Histopathology, Olabisi Onabanjo University, Nigeria
Correspondence: Mohammad Ilyas, Division of cancer and Stem Cell, Faculty of Medicine and Health Sciences, University of Nottingham, Nottingham, UK
Received: October 31, 2016 | Published: March 7, 2017
Citation: Ham-Karim HA, Ebili HO, Fadhil W, et al. COLD-HRM: a combination of methods to infer the nature of somatic mutations. Adv Cytol Pathol. 2017;2(2):54–61. DOI: 10.15406/acp.2017.02.00017
High Resolution Melting (HRM) measures DNA melting patterns to detect mutations. Hitherto, HRM has not been used to report the nature of the mutation. We reasoned that in conditions with a minor allele (e.g. allelic loss and somatic mutation), CO-amplification at Lower Denaturation temperature (COLD) PCR could change HRM patterns allowing (i) detection of Loss of Heterozygosity (LOH) and (ii) discrimination of somatic mutations from germline variants.
For LOH, we tested paired tumour/normal samples from 11 cases of colorectal cancer (CRC) which were heterozygous for the SMAD4 SNP rs12455792 and 1 case which was homozygous wild type. To discriminate somatic from germline mutation, we tested tumour from 4 CRCs with somatic mutation in codon 374 POLE, paired tumour/normal tissue from 10 CRCs heterozygous for POLE SNP rs4077170 and matched normal samples wild type. All samples underwent both the standard and COLD PCR followed by HRM analysis. In 4/11 tumour samples which were heterozygous for the SMAD4 SNP, HRM patterns changed following COLD-PCR suggesting LOH. Changes in HRM patterns were also seen in 4/4 tumours with POLE codon 374 mutation suggesting somatic rather than germline mutation. No change was seen in the 20 tumour and matched normal samples heterozygous for POLE SNP rs4077170. We conclude that combining COLD PCR with HRM (COLD-HRM) allows inference of LOH and somatic sequence mutation without the requirement for matched normal DNA.
Keywords: COLD-PCR, HRM, loss of heterozygosity, somatic mutation, germline mutation, single nucleotide polymorphism
COLD PCR, CO-amplification at lower denaturation temperature PCR; LOH, loss of heterozygosity; HRM, high resolution melting; SSCP, single strand conformation analysis
Loss of Heterozygosity (LOH) and sequence mutation are two frequently occurring types of somatic alterations in cancer.1-3 Detection of these changes is important as it delineates the profile of a cancer and it may also provide prognostic and predictive information.4-12 A variety of techniques exist for detection of these mutations. LOH can be detected, at the gross chromosomal level, with techniques such as array-based chromosomal genomic hybridization (aCGH), karyotyping and fluorescent in-situ hybridization (FISH).1,7-9 For a higher resolution analysis, genotyping through quantification of Single Nucleotide Polymorphisms (SNPs) or microsatellites can be performed.13-15 These methods of detecting LOH can be cumbersome and often require matching non-tumour DNA from the same patient.1,7,13-15
The gold standard for detecting somatic sequence mutations is DNA sequencing.16 However, unless there is a well-defined and narrow mutation hot-spot (such as mutations of codon 12/13 in KRAS), large areas of DNA may need to the tested costing both time and money. Costs can be reduced by screening the DNA for the possible presence of mutations using techniques such as High Resolution Melting (HRM), Single Strand Conformation Analysis (SSCP) and denaturing High Performance Liquid Chromatography (dHPLC).17-19 Such methods interrogate the physical characteristics of the DNA which are altered by a change in the DNA sequence. These methods cannot however distinguish between germline SNPs and cancer-specific somatic changes unless normal DNA from the patient is available for comparison.
The currently available techniques for detecting LOH and somatic sequence mutations can be time- consuming and expensive and an improved methodology-in particular one which would remove the requirements for samples of matched normal DNA-is required. In this paper, we report developments to achieve this. Our method depends firstly on the fact that both LOH and sequence mutation induce a state of major and minor alleles of the target gene1-3 in a tumour. LOH forces a minority state on the wild type allele due to loss of chromatin or gene conversion whilst, in the case of somatic sequence mutation, the presence of contaminating wild type DNA from stromal cells, will result in the mutant allele becoming a minor allele.1-3 Secondly, we used the fact that the frequency of the minor allele can change due to the minor allele-enrichment capability of the Co-Amplification at Lower Denaturation temperature (COLD) PCR protocol.3 In contrast, when there is no minor allele (for example a heterozygous SNP), there will be no change in allele following COLD-PCR. Finally, our method depends on the fact that HRM data can be used to detect the changes in minor allele frequency.3,20-24
HRM is a simple and robust method for detection of sequence mutations such as point mutations, small indels (including microsatellite instability), and gene promoter methylation (following bisulphite treatment).17,18,25-19 The underlying principle is that mutant and wild-type DNA sequences can be pushed into forming heteroduplexes which melt differently to homoduplexes.15,16 When testing the limit of detection of the HRM methodology in a previous study, we noticed that melting patterns were influenced by the proportion of mutant alleles.27 We reasoned that any change in mutant allele frequency should be detectable by a change in melting pattern and thus any sample which contained a minor allele in a particular gene would show a change in melting pattern following COLD-PCR. Our hypothesis therefore was that comparing the melting pattern of the same DNA sample following standard and full COLD-PCR would demonstrate the presence of a minor allele. This in turn would allow us to infer the presence of LOH or somatic sequence mutation (rather than a germline SNP) without the need for a matched normal sample.
Mutant allele quantification by High Resolution Melting (HRM) analysis In order to confirm the quantitative nature of the melting pattern in HRM, we used the colorectal cancer cell lines HCT116 and Vaco5 (both kindly donated by professor Ian Tomlinson). HCT116 contains a heterozygous KRAS G13D mutation whilst Vaco5 is wild type for KRAS. Variant quantities of DNA from the cell lines were mixed to produce mixtures containing KRAS G13D mutant alleles at a frequency of 25%, 12.5%, 6.25%, 3.75%, 1.5% and 0.75%. These mixtures were subjected to COLD PCR (primer sequences and some cycling conditions are indicated in Tables 1 & 2) followed by HRM analysis (see below).
Gene Region |
Forward Primer |
Reverse Primer |
KRAS codon 13 |
5’-TGAATATAAACTTGTGGTAGTTGG-3’ |
5’-GCTGTATCGTCAAGGCACTCT-3’ |
SMAD4 rs12455792 |
5`-GCCATTTTGTAATCCCAGCA-3’ |
5`-GCACATGAAAGATGCTCGAC-3’ |
POLE codon 374 |
5’-TGTCTGTCCTCTTTCCAACC-3’ |
5’-AGCCATGCTGCTCTGTGG-3’ |
POLE rs4077170 |
5’-CCTCTGACTTGTGCTGATTGC-3’ |
5’-CCGGGTAGTTTCCCAAGTG-3’ |
Table 1 List of primer pairs used in the COLD HRM experiments.
Parameters |
Temperature (0C) |
Duration |
Number of Cycle |
Initial Denaturation |
95 |
5 min |
1 |
Denaturation |
95 |
10 sec |
45 |
Heteroduplex forming temperature |
70 |
20 sec |
|
Critical temperature Tc |
85.3 (SMAD4) |
10 sec |
|
82.3 (POLE* & KRAScodon 13) |
|||
Annealing |
55 (SMAD4 & POLE*) |
45 sec |
|
60 (KRAS codon 13) |
|||
Extension |
72 |
20 |
|
Final extension |
72 |
5 min |
1 |
Table 2 Cycling parameters for COLD PCR.
*POLE: rs4077170 and codon 374.
Case selection Approval of the study and access to the tissue were granted by Nottingham Health Sciences Biobank. All experiments to test for LOH and somatic sequence change were performed on DNA obtained from formalin-fixed, paraffin-embedded (FFPE) tissue samples and all selected for testing were identified from a previous unpublished study. In order to test for LOH, we decided to interrogate a SNP at the SMAD4 locus on chromosome 18q-a common site of allelic loss in colorectal cancer. Eleven cases with heterozygosity in the germline for this SNP and 1 case which were homozygous for the wild type sequence at this locus were selected. Tumour blocks and matched normal blocks were retrieved from the pathology archives of the Nottingham University Hospitals of pathology department.
In order to discriminate germline sequence variation from somatic mutation, we tested 4 cases of CRC with known mutations in codon 374 of the POLE gene. As a comparison for these cases, we also tested 10 cases (tumour and matched normal) with a known germline SNP at POLE (rs4077170) but without somatic mutation. Ten (10) samples of normal tissue which were homozygous at this locus were used as controls.
Polymerase chain reaction All PCRs were performed on the Primus 96 Advanced Gradient Thermal Cycler in a 10µl volume made up of 5µl of Diamond Hotshot master mix (Clent Life Science), 250nm primer (Eurofins) concentrations, 1µl of LC Green (Idaho Technology/Biofire Diagnostics) and 20ng of DNA template. The SNP, located in the intronic region of SMAD4 on chromosome 18, is a C>T polymorphism, the C allele being the wild type and the T the variant. The global minor allele frequency is 0.3746 (37.461%), making it a common polymorphism and thus suitable for single gene LOH analysis. The POLE SNP rs4077170 (G>C) is similarly intronic and a common variant with a minor allele frequency of 0.3179 (31.79%) to 0.4283 (42.83%).
The primer sequences and standard PCR thermal cycling conditions for rs12455792, rs4077170 and exon12 of POLE (containing codon 374) are given in Tables 1 & 3. Primer sequences were designed using Primer 3. In order to perform full COLD-PCR, it is essential to ascertain the critical temperature (Tc) at which heteroduplexes of mutant/wild type allele will melt whilst homoduplexes will remain double stranded. We identified the Tc for each of the PCR reactions using a gradient PCR machine as previously described.3,20,21 The cycling conditions for full COLD-PCR are given in Table 2.
Parameters |
Temperature (0C) |
Duration |
Number of Cycle |
Initial Denaturation |
95 |
5 mins |
1 |
Denaturation |
95 |
10 sec |
40 (POLE)* |
Annealing |
55 |
30 sec (POLE)* |
|
45 sec (SMAD4) |
|||
Extension |
72 |
20 |
|
Final extension |
72 |
5 mins |
1 |
Table 3 Cycling parameters for standard PCR.
*POLE: rs4077170 and codon 374.
High resolution melting and analysis Melting was performed on the Light Scanner-96 platform (Idaho Technology/ Biofire Diagnostics) using the following melting parameters. The temperature range was set at 650C – 950C and to ‘Auto’ exposure and a ramp rate of 0.10C/sec. Sample equilibration was performed at 620C. The high resolution CCD camera captured the fluorescence data for analyses. Thereafter, analyses were performed with the Light Scanner Call-IT software version 2.0.0.1.331 using the Expert scanning module. The negative control samples as well as the poorly amplified samples were excluded from further analyses using the negative filter. Normalization of the melting data was done as per manufacturer’s instructions. The normalized melting curves were shifted at a 0.05 level, while sensitivity was set to normal at zero level. The internal standard, ‘Auto group’ and ‘Common vs Variant’ functions were used to group the normalized and shifted melting curves according to their melting patterns. The results were viewed in the ‘Shifted melt curve’ and ‘Difference curves’ outputs.
Sequencing The sequencing PCRs were performed in 25 µl reactions and purified using the QI Aquick kit (Qiagen). The products were sequenced using Applied Biosystems Big Dye Terminator v3.1 Cycle Sequencing Kit and 3130 xl Genetic Analyzer. The electropherograms were analysed using Finch TV software version 2.01.
HRM is quantitative DNA samples with varying ratios of mutant and wild type DNA for KRAS codon 13 were tested by PCR followed by HRM. The Difference curve of the HRM analysis shows the change in fluorescence of a test sample compared with a control which consists only of homoduplexes. The data showed that the melting patterns between the samples were different and that the greater the proportion of mutant DNA (and therefore a higher proportion of heteroduplexes), the greater the difference in fluorescence between test sample and control. The differences were not linearly related to the mutant allele proportion and cannot therefore be used to precisely quantify mutant alleles. These results do however show that HRM can roughly quantify the proportion of variant DNA in a mixture with wild type template (Figure 1).
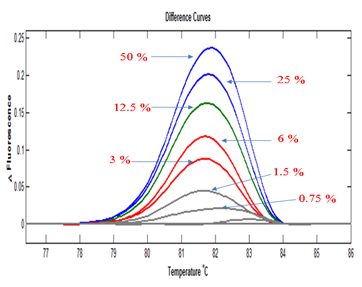
Figure 1 Test of quantitative capability of HRMA. HRMA can separate samples on the basis of proportion of variant allele they contain. The sample containing 50% mutant is the un-mixed DNA from the KRAS-codon-13 heterozygous HCT116 cell line.
COLD-HRM and LOH DNA samples from 12 cases of CRC and matched normal tissue underwent standard and full COLD-PCRs amplification. Eleven (11) of these cases were known to have rs12455792 SNP at SMAD4, while 1 case was homozygous wild type at this locus. The PCR products were analysed by HRM and the melting patterns compared. In 4/11 cases of CRC with the SNP, there was a difference in the melting pattern between the COLD-PCR-amplified and the standard PCR-amplified products. In each case this was characterised by an increase in the height of the peak COLD-PCR amplification products over those of the standard PCR in the Difference plot (Figures 2A - 2D). This corresponds to an enrichment of the minor allele following COLD-PCR process suggesting allelic imbalance at the rs12455792 SNP in SMAD4. Seven of the 11(7/11) cases did not show this differential melting pattern (data not shown).
Direct sequence analysis was performed for the 4 cases with differential melting patterns between the standard PCR and COLD-PCR. The results showed an increase in the height of the wild type C allele peak in 3 cases and an increase in the height of the variant T allele peak in the remaining case. This was confirmation that the ratio of the alleles had changed and that the minor allele had increased in frequency, hence the differential melting pattern between standard PCR- and COLD PCR-amplified products (Figures 2A - 2D). The matched normal tissue for these four cases showed no change in the melting pattern following COLD-PCR (only one shown in Figure 2E: sample 5).
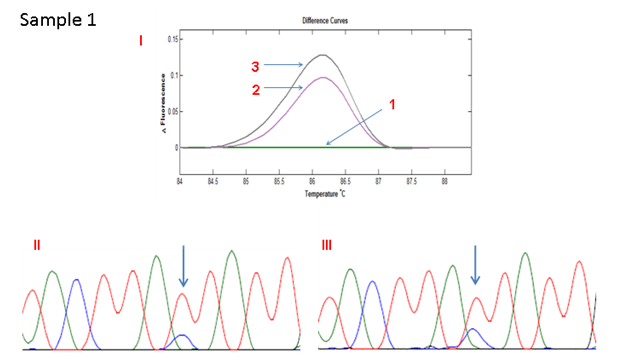
Figure 2A HRMA of the standard and full COLD PCR products of SMAD4 rs12455792 locus in homozygous wild type reference and tumour DNA showing enrichment of the minor allele by COLD PCR in the tumour DNA, a confirmation of LOH in the tumour DNA at rs12455792. (I) 1: control without SNP; 2: Tumour with SNP (amplified by standard PCR); 3: Tumour with SNP (amplified by COLD PCR). (II) Sanger sequencing of tumour sample (amplified by standard PCR). (III) Sanger sequencing of tumour sample (amplified by COLD PCR).
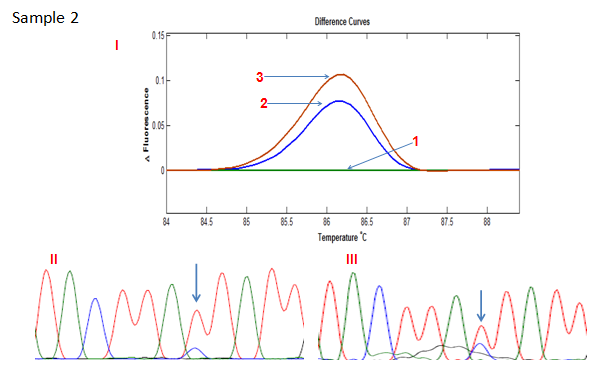
Figure 2B HRMA of the standard and full COLD PCR products of SMAD4 rs12455792 locus in homozygous wild type reference and tumour DNA showing enrichment of the minor allele by COLD PCR in the tumour DNA, a confirmation of LOH in the tumour DNA at rs12455792. (I) 1: control without SNP; 2: Tumour with SNP (amplified by standard PCR); 3: Tumour with SNP (amplified by COLD PCR). (II) Sanger sequencing of tumour sample (amplified by standard PCR). (III) Sanger sequencing of tumour sample (amplified by COLD PCR).
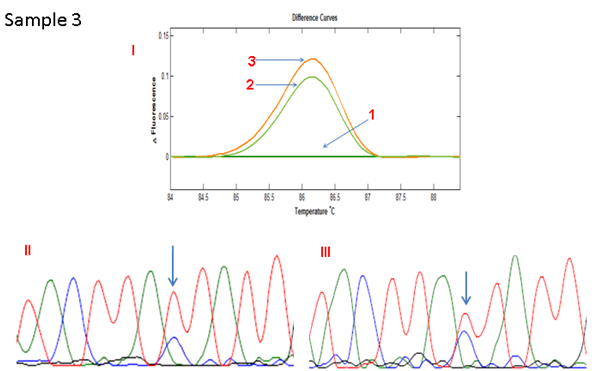
Figure 2C HRMA of the standard and full COLD PCR products of SMAD4 rs12455792 locus in homozygous wild-type reference and tumour DNA showing enrichment of the minor allele by COLD PCR in the tumour DNA, a confirmation of LOH in the tumour DNA at rs12455792. (I) 1: control without SNP; 2: normal with SNP (standard and COLD PCR); 3: Tumour with SNP (amplified by standard PCR); 4: Tumour with SNP (amplified by COLD PCR). (II) Sanger sequencing of tumour sample (amplified by standard PCR). (III) Sanger sequencing of tumour sample (amplified by COLD PCR).
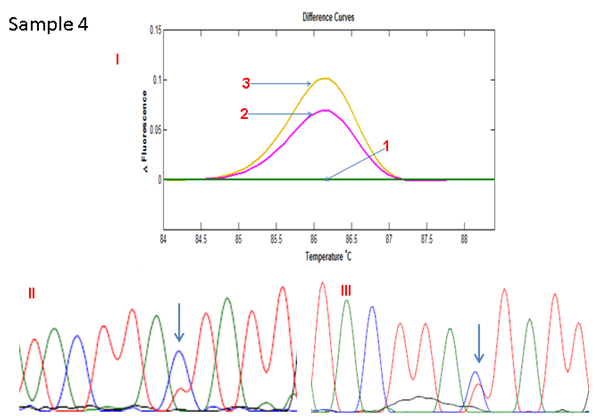
Figure 2D HRMA of the standard and full COLD PCR products of SMAD4 rs12455792 locus in homozygous wild-type reference and tumour DNA showing enrichment of the minor allele by COLD PCR in the tumour DNA, a confirmation of LOH in the tumour DNA at rs12455792. (I) 1: control without SNP; 2: Tumour with SNP (amplified by standard PCR); 3: Tumour with SNP (amplified by COLD PCR). (II) Sanger sequencing of tumour sample (amplified by standard PCR). (III) Sanger sequencing of tumour sample (amplified by COLD PCR).
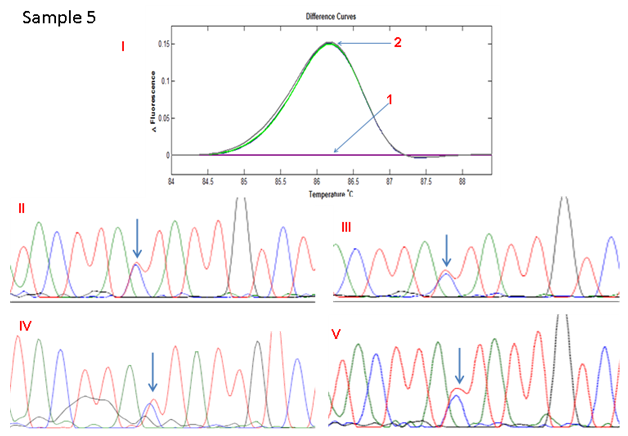
Figure 2E HRMA of the standard and full COLD PCR products of SMAD4 rs12455792 locus in homozygous wild type reference and tumour DNA showing no change in heights of peaks of standard and COLD PCR-amplified products in the tumour, suggestive of allelic balance at rs12455792 in the tumour DNA.(I)1: control without SNP; 2: standard and Cold-PCR products of tumor with SNP; (II): Sanger sequencing of wild type reference (amplified by Standard PCR), III: Sanger sequencing of wild type reference (amplified by COLD PCR), IV: Sanger sequencing of tumour (amplified by Standard PCR), V: Sanger sequencing of tumour (amplified by COLD PCR).
COLD-HRM and somatic sequence mutation Tumour tissue from 4 cases of CRC with a known mutation in codon 374 of POLE underwent analysis by standard PCR and COLD-PCR amplification followed by HRM. Comparison of the data showed that in all of these cases, there were differences in the melting pattern between standard PCR- and COLD-PCR-amplified tumour samples in every case (Figures 3A & 3B). As expected, there was an increase in the size of the peak following COLD-PCR which indicates enrichment of the minor allele. This was confirmed by DNA sequencing which showed an increase in the signal from the mutant allele in all 4 cases (Figures 3C & 3D).
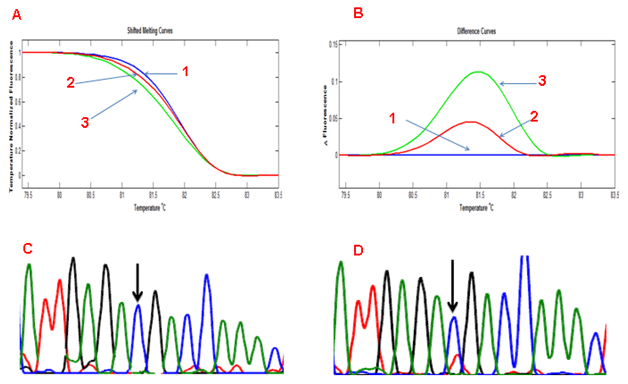
Figure 3 HRMA of a standard PCR- and COLD PCR-amplified sample with somatic mutation at POL E codon 374 showing (A) a left shift of the COLD PCR-amplified product (Shifted Melting Curves) and (B) a rise in the curve peak on the Difference Curves. 1: wild type sample, 2: Tumour (standard PCR), 3: Tumour (COLD PCR). (C) Sanger sequencing of tumour sample (amplified by standard PCR). (D) Sanger sequencing of tumour sample (amplified by Cold-PCR).
Our hypothesis is based on the prediction that a minor allele (whether due to somatic mutation or LOH at a SNP locus) will undergo enrichment with COLD-PCR thus altering the HRM pattern whilst a heterozygous allele (at a frequency of 50%) will remain unchanged. The lack of change was seen in the experiments with the SNP in SMAD4 but, in order to exclude a locus-specific effect, we sought to validate this using a different SNP. The POLE SNP rs4077170 (G>C) was tested in a series of 10 cases of matched tumour and normal. In all 20 samples, the melting pattern was identical whether standard PCR or COLD-PCR was performed (Figures 4A & 4B). Sequencing of 10 of the cases (5 tumour and 5 normal samples) similarly showed that there was no change in the ratio of alleles (Figures 4C - 4F: only 1 normal and 1 tumour samples shown).
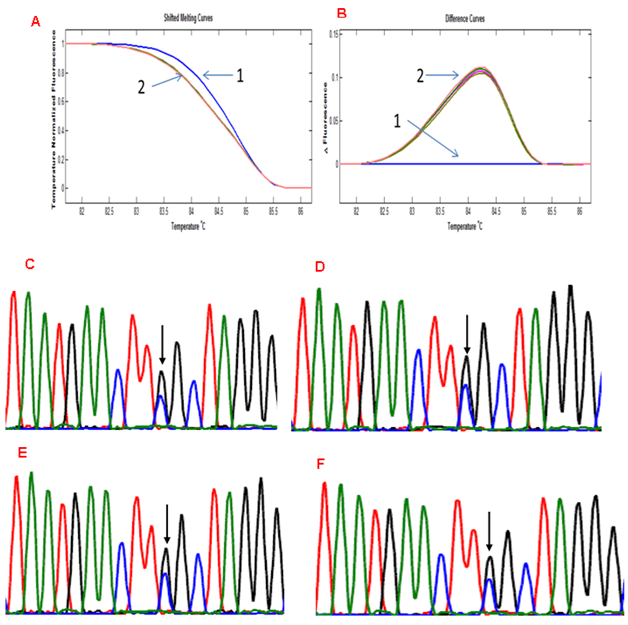
Figure 4 HRMA of tumour and matched normal sample having the rs4077170 SNP at POLE showing no significant change in melting pattern between standard PCR- and COLD PCR-amplified products. 1: Normal without SNP, 2: matched normal and tumour with the rs4077170 SNP. Electropherogram of 2 samples with POLE rs4077170 SNP. No change is seen in the peak of any of the alleles between standard PCR- and COLD PCR-amplified products. C: Tumour (Standard PCR), D: Tumour (COLD PCR), E: Normal (Standard PCR), F: Normal COLD PCR).
In this study we have sought to combine two methodologies in order to facilitate mutation analysis of tumours. We aimed to establish a simple protocol which will (i) allow LOH to be evaluated in tumour samples, (ii) discriminate somatic sequence mutations from germline SNPs and (iii) remove the requirement of matched normal DNA samples. Our protocol was based on the fact that both LOH and somatic sequence mutation create minor allele which can be enriched by COLD-PCR. This enrichment can be detected by HRM since the data are quantitative. COLD-HRM is a previously coined portmanteau term which acknowledges the contribution from both methods although it was used for a different purpose.29
HRM has previously been shown to be quantitative22-24 and this was confirmed in our initial experiments. These showed that the proportion of the minor allele can affect the pattern of DNA melting resulting in peaks whose size correlates with the proportion of the minor allele. As the correlation is not linear, it cannot be used to precisely quantify the minor allele but it can be used to discriminate between samples. For both evaluation of LOH and somatic sequence mutation, our methodology was based on the assumption that heterozygous SNPs would remain in a 50/50 ratio following COLD-PCR since there was no minor allele. In order to confirm this we tested 36 samples (including both normal tissue and tumour tissue) at 3 different loci. In all cases, both standard and COLD PCR were performed, and PCR results were validated by sequencing analysis in some of these cases.
For the analysis of LOH, we tested 12 CRCs, 11 of which were known to have a SNP in the SMAD4 gene. In 4 cases (36.3%) there was a difference in melting pattern between standard PCR and COLD-PCR. Minor allele enrichment was confirmed by sequencing and thereby validating this method as a robust means of detecting LOH. This method is quicker than FISH and, since it does not require normal matched DNA samples, it is more flexible than testing for LOH using microsatellites. It is also quicker than massively parallel technologies (such as microarrays or next generation sequencing) and its role would be more in low throughput targeted research or diagnostics. In addition, it would have a role in laboratories where the budget is limited. Since LOH is a form of allelic imbalance, it is conceivable that the same principle could be used to test for gene amplification if a SNP is identified within the amplified region. In this case the non-amplified allele may be the minor allele. For somatic mutation detection, 4 cases of CRC known to have a mutation in codon 374 of POLE were tested. There was mutant allele enrichment following COLD-PCR which was validated by sequencing in all 4 cases.24,29 Our data confirm that COLD-HRM can be used to discriminate somatic sequence mutations from germline SNPs in tumours without the need to compare with matched normal DNA.
The method we have described is simple and robust but it does have some caveats. Firstly, the PCR has to be done twice i.e. a standard protocol and the COLD-PCR protocol and this requires the identification of the Tc for each target. This is however outweighed by the advantage of not requiring normal DNA for each tumour. It is possible to calculate the Tc using software which can predict the melting temperature of heteroduplexes therefore removing the need to individually test each target.30 Another caveat is that this method depends on the presence of a minor allele. In most cases, a mutant allele will be the minor allele due to the presence of stroma within a tumour. It is possible that, if there is a high proportion of tumour cells and there are events such as copy-neutral LOH, the frequency of the mutant allele approaches 50%. This may give a false negative result although it is anticipated that this will not occur often. In summary, we have the combined methodologies to improve mutation detection in tumours.
We have proposed a refined interpretation of COLD-HRM which allows inference of LOH and somatic sequence mutation based on presence or absence of melting pattern following COLD-PCR. This protocol is cheap, quick and robust and, most importantly, it does not require matched normal DNA.
The authors wish to thank the other members of the Pathology Research Group, for the technical support provided in the development of this methology.
The author declares no conflict of interest.

©2017 Ham-Karim, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.