MOJ
eISSN: 2379-6294


Research Article Volume 1 Issue 5
1Department of Forensic Medicine and Clinical Toxicology, Cairo University, Egypt
2Department of Biochemistry, Cairo University, Egypt
3Department of Andrology, Cairo University, Egypt
Correspondence: Lamis M Nader, Department of Forensic Medicine and Clinical Toxicology, Faculty of Medicine, Cairo University, Egypt, Tel 9715 0267 1573
Received: May 13, 2015 | Published: December 9, 2015
Citation: Nader LM, Mohamed AS, Ismail MM, et al. Molecular biomarkers: the development of mrna multiplex rt-pcr assay for the definitive identification of semen. MOJ Toxicol. 2015;1(5):169–177. DOI: 10.15406/mojt.2015.01.00025
Background: Semen is one of the most common body fluids found at the crime scene. Protamine-1 (PRM1), protamine-2 (PRM2) and semenogelin-1 (SEMG1) are specific mRNA markers found only in semen so if it is possible to detect these markers in a forensic specimen by RT-PCR, this could be helpful to verify the presence of semen.
Aim of the work: Identification of semen in forensic samples by molecular assessment of specific RNA biomarkers.
Subjects and methods: Semen samples were obtained from 50 inpatients and outpatients of the andrology department that were divided into 3 groups including: normozoospermic (21 specimen), oligozoospermic (11 specimen) and azoospermic (18 specimen) groups and 10 blood samples were taken from the normozoospermic patients to assess the presence of the markers in blood. The next steps of work were RNA extraction from semen and blood, PCR amplification of genes, detection of the amplified gene using agarose gel electrophoresis and performing gene expression analysis of the 3 genes using quantitative RT-PCR.
Results: PRM1, PRM2 and SEMG1 were only detected in semen samples and were totally absent in blood samples. PRM1 was the most specific and reliable marker for semen identification followed by PRM2 and SEMG1 which was the least specific. These markers are not only used for identification but also for detection of the semen profile of the There was a direct positive correlation between sperm count and RNA expression.
Conclusion: The analysis of the RNA profile in a sample can uniquely identify the fluid or tissue of origin. RT- PCR is valuable in semen identification by gene expression analysis of PRM1, PRM2 and SEMG1. PRM1 was the most specific and reliable marker used for semen identification.
Keywords: semen identification, rt-pcr, protamine-1, protamine-2, semenogelin-1
PRM1, protamine-1; PRM2, protamine-2; SEMG1, semenogelin-1; PSA, prostate-specific antigen; RT-PCR, reverse transcriptase-polymerase chain reaction
Determination of the type and origin of biological samples found at crime scenes can provide important clues for crime scene reconstructions by supporting a link between sample donors and actual criminal acts. The presence of certain body fluids can be used as excellent indicators of the sequence of events which occurred. For example, blood stains can indicate some form of physical struggle, assault or murder, and detection of semen or vaginal fluid can indicate the involvement of some form of sexual assault. The common body fluids found at crime scenes are blood, semen, saliva, vaginal fluid, urine, and sweat.1
The first step of identifying a particular body fluid is highly important since the nature of the fluid is itself very informative to the investigation, and the destructive nature of a screening test must be considered when only a small amount of material is available. The ability to characterize an unknown stain at the scene of the crime without having to wait for results from a laboratory is another very critical step in the development of forensic body fluid analysis. Driven by the importance for forensic applications, body fluid identification methods have been extensively developed in recent years.2
Semen is one of the most commonly encountered body fluids at crime scenes. The total volume of semen for each ejaculation of human male varies from 2 and 5ml3 (0.12 to 0.31 cubic inch). In human beings each ejaculation contains normally 200 to 300 million sperm, making semen ideal for DNA profiling.3
Semen can be detected using an ultraviolet light. It is a routine procedure to search for semen and other fluids using this simple and non-destructive method.4 The most popular confirmatory test for semen beyond looking for sperm cells is the test for prostate-specific antigen (PSA). The semen from azoospermic males will still contain this antigen which is present in the seminal plasma; other body fluids contain a very low level of PSA and do not interfere. Many of the new methods being developed to identify semen stains involve the detection of specific messenger ribonucleic acid (mRNA) markers .5
The method of biological fluid identification through mRNA profiling takes advantage of the fact that cells present in various biological fluids show specific patterns of gene expression and they synthesize proteins in characteristic amounts. The method is very sensitive owing to the use of amplification (PCR) and is also specific thanks to the unique pattern of gene expression for cells and organs with various functions. There are different mRNA markers that are frequently used in forensic science in order to determine the nature of forensic samples. Protamine-1 (PRM1), protamine-2 (PRM2) and semenogelin-1 (SEMG1) are specific RNA markers for semen identification. Protamine-1 and protamine-2, two sperm markers, encode protamine which substitutes histones in spermatozoa and one seminal plasma marker encodes a semenogelin an abundant protein in seminal fluid involved in the gelatinous entrapment of ejaculated spermatozoa.6
Sakurada et al.7 demonstrated the feasibility of identifying body fluids by using a real-time PCR assay (RT-PCR). In RT-PCR, mRNA is transcribed into DNA (complementary DNA, cDNA) using the reverse transcriptase (RT enzyme) and a transcript-specific primer.8 The cDNA is amplified using primers binding to the target sequence of interest. Real-time PCR and real-time reverse transcription PCR have catalyzed dramatic changes in many disciplines of biology. RT-PCR is the technique that allows collection of data throughout the process of PCR. Real-time PCR is capable of combining amplification and detection into a single step .9
Real-time PCR allows more quantitative determination of the transcript, which may be of significance when certain markers appear in fluids other than theoretically specific for a given marker due to contaminations, e.g., haemoglobin A (HBA) in saliva in bleeding gums or mucin 4 (MUC4) in sperm if the donor has recently had sexual intercourse.10
The present study aimed for the identification of semen from other body fluids in forensic samples by molecular assessment of specific RNA biomarkers.
This work was performed with the cooperation of the Forensic Medicine and Clinical Toxicology Department, Molecular Biology Unit, Medical Biochemistry Department and Andrology Department, Faculty of Medicine, Cairo University.
Sample collection
A total of 50 males were recruited from the outpatients attending the Andrology clinic & inpatients of Andrology department of Cairo University hospital after approval and signed informed consents IRB. All of them had been married for more than 2 years.
Exclusion criteria were smoking and leucocytospermia. A detailed medical history was taken and physical examination was performed for the investigated cases.
Semen ejaculates were obtained after 4 days of sexual abstinence. The samples were examined immediately after liquefaction according to WHO guidelines (1999) (normal sperm count >20 X 106 sperm ml-1, sperm motility >50%, abnormal sperm morphology <70% and leucocytes <106 ml-1).
Semen samples were classified according to their sperm count into three groups:
Blood samples were collected on anticoagulant vacationer tubes by venipuncture from only 10 of the control subjects after informing them all and getting their consent. Semen samples were stored at -80°C and blood samples were subjected to RBCs lysis buffer and stored at -4°C until performing the RNA extraction.
Extraction of RNA from the blood cells and seminal body fluids
Total RNA was extracted from collected samples using Total RNA Isolation system (QIAGEN, CA, Germany). The yield of total RNA obtained was determined spectrophotometrically at 260 nm.
Primer design and selection
All primers were designed based on target sequences obtained from the references. For the selection of the ideal primer pair, the considered factors included melting temperature (Tm: 60-65°C), GC content (40%-60%), and amplicon length of about 90-200bp.
Sequence of primers as following (Table 1):
|
Forward Primer |
Reverse Primer |
Protamine-1 |
5′-GCACCATGGCCAGGTACAGATGC-3′ |
5′-GCATTGTTCCTTAGCAGGCTCCTG-3′ |
Protamine-2 |
5'-GGATCCACAGCCGCCAGCATCGCT-3' |
5'- GCATGTTCTCTTCCTGGTTCTGCA-3' |
Semenogelin-1 |
5′-GCG GTA CCC ATG AAG GGA AAC TCA CAT TT-3′ |
5′-GCA GAT CTT GAG AGC TGA GCC GAC CTT GT-3′ |
Table 1 Sequence of primers
Calculation of relative quantification (RQ) (relative expression)
After the RT-PCR run the data were expressed in Cycle threshold (Ct). The PCR data sheet includes Ct values of assessed gene (VEGF-R2 and eNOS) and the house keeping (reference) gene- the gene that continuously and normally expressed in the cell- (GAPDH). In order to measure the gene expression of certain gene, a -ve control (reference) sample should be used. Therefore, target gene expression was assessed and related to reference (internal control) gene as follows:
Finally, RQ was calculated according to the following equation:
Detection of the amplified DNA using agarose gel electrophoresis
At the end of the amplification process, DNA agarose gel electrophoresis was used to detect the presence of the gene. The DNA product was visualized by placing the gel on an UV transilluminator. The EB intercalated into DNA and gave a bright pink band.12
PCR product yielded a 333bp fragment on 1.5% agarose gel electrophoresis using 100bp DNA ladder (Jena Bioscience, Germany) as recorded by the gel documentation system.

The data were coded and analyzed using the statistical package SPSS version 17. The data were summarized using descriptive statistics: mean, standard deviation, median, minimal and maximum values for quantitative variables. Statistical differences between groups were tested using ANOVA (analysis of variance) with post Hoc Bonferroni test for quantitative normally distributed variables while nonparametric Mann Whitney test and kruskal-Wallis test were used for quantitative variables which weren’t normally distributed.
Wilcoxon Signed Ranks Test was used for pair wise comparison. Correlations were done to test for linear relations between variables. Logistic regression analysis was done to test for significant predictors of azospermia and/or oligospermia. P-values £ 0.05 were considered statistically significant .13
In the present work, it was proposed to analyze semen samples for detection of protamine-1 (PRM1), protamine-2 (PRM2) and semengolin-1 (SEMG1) as indicators of the presence of semen and differentiating it from blood of the same person as an example of body fluid.
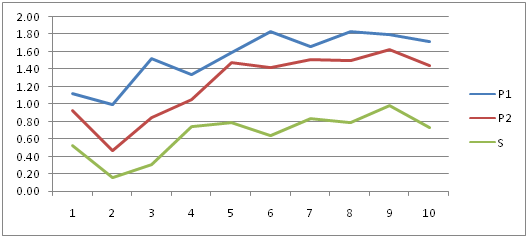
Figure 1 showing the Cycle threshold (Ct) values of RNA expression in PRM1, PRM2 and SEMG1 in normozoospermic group. It is clear that PRM1 has got the highest levels of expression so it was the most specific and reliable marker for semen identification. PRM2 was the next in specificity followed by SEMG1.
Table 2 summarized the descriptive data of the studied subjects in the present study. It presented the mean ± SD, median, minimum, and maximum of the subject’s age in years, sperm count/ml in million, motility and abnormal forms as well as PRM1, PRM2 and SEMG1 levels measured by RT-PCR in the studied 50 samples.
Studied Parameter |
Mean ±SD |
Median |
Minimum |
Maximum |
Age (year) |
36.12 ± 7.63 |
33.5 |
26 |
50 |
Count /ml (million) |
39.50±45.35 |
8.5 |
0 |
123 |
Motility |
29.36 ± 27.50 |
25 |
0 |
70 |
Abnormal forms |
20.60 ±17.95 |
25 |
0 |
63 |
PRM1 |
0.85±0.55 |
0.82 |
0 |
1.83 |
PRM2 |
0.73±0.42 |
0.69 |
0 |
1.63 |
SEMG1 |
0.42±0.25 |
0.36 |
0 |
0.98 |
Table 2 Descriptive data of the studied subjects
All data were expressed as mean ±SD (standard deviation)
P value was considered significant at (p≤ 0.05)
PRM1=Protamine-1, PRM2=Protamine-2 and SEMG1 =Semenogelin-1
Table 3 showed the sperm count/ml (million), motility, abnormal forms, PRM1, PRM2 and SEMG1 levels in the normozoospermic, oligozoospermic and azoospermic groups. There was a statistical significant difference between the different groups concerning sperm count/ml in million, motility, abnormal forms as well as PRM1, PRM2 and SEMG1 levels (p< 0.001** for all).
Semen Analysis |
Normozoospermic Group (21 Samples) |
Oligozoospermic Group (11 samples) |
Azoospermic Group (18 samples) |
Kruskal Wallis test |
Count / ml (million) |
87.48±27.30 |
12.55±12.02 |
0.00±0.00 |
0.00** |
Motility |
58.00±9.87 |
22.73±13.67 |
0.00±0.00 |
0.00** |
Abnormal forms |
31.90±10.42 |
32.73±12.92 |
0.00±0.00 |
0.00** |
PRM1 |
1.37±0.35 |
0.58 ± 0.30 |
0.40±0.29 |
0.00** |
PRM2 |
1.11± 0.35 |
0.53 ±0.17 |
0.42±0.21 |
0.00** |
SEMG1 |
0.62 ± 0.24 |
0.29 ±0.13 |
0.26±0.14 |
0.00** |
Table 3 The sperm count, motility, abnormal forms, PRM1, PRM2 and SEMG1 in each studied group
All data were expressed as mean ±SD (standard deviation)
PRM1=Protamine-1, PRM2=Protamine-2 and SEMG1 =Semenogelin-1
**Statistical significance was set at p≤ 0.05
Table 4 showed the Mann-Whitney test results which were used to compare the different studied groups. It revealed a statistical significant difference existed between azoospermic and oligozoospermic groups concerning sperm count/ml in million, motility, abnormal forms (p<0.001**) with no statistical difference in PRM1, PRM2 and SEMG1 levels between these 2 groups. Similarly, a statistical significant difference existed between azoospermic and normozoospermic groups as well as between oligozoospermic and normozoospermic groups concerning sperm count/ml in million, motility, abnormal forms, PRM1, PRM2 and SEMG1 levels (p<0.001** for all).
Compared |
Count /ml |
Motility |
Abnormal |
PRM1 |
PRM2 |
SEMG1 |
Azoospermic & |
0.000** |
0.000** |
0.000** |
0.120 |
0.251 |
0.589 |
Azoospermic & |
0.000** |
0.000** |
0.000** |
0.000** |
0.000** |
0.000** |
Oligozoospermic & |
0.000** |
0.000** |
0.133 |
0.000** |
0.000** |
0.001** |
Table 4 Mann-Whitney test comparing the different studied groups
**Statistical significance was set at p≤0.05
PRM1=Protamine-1, PRM2=Protamine-2 and SEMG1 =Semenogelin-1
Table 5 & Figure 2-7 presented the correlations studies between the different investigated parameters. It showed a statistical significant positive correlation between age and PRM1 and SEMG1 levels (r=0.324 and p=0.022* and r=0.309 and p=0.029* respectively).
Spearman's Rho |
PRM1 |
PRM2 |
SEMG1 |
|
|
AGE Correlation Coefficient (r)
|
0.324 |
0.252 |
0.309 |
Count/ml (million)Correlation Coefficient (r) |
0.842 |
0.810 |
0.687 |
|
Motility Correlation Coefficient (r) |
0.786 |
0.742 |
0.624* |
|
Abnormal forms Correlation Coefficient (r) |
0.489 |
0.502 |
0.380 |
|
PRM1 Correlation Coefficient (r) |
1.000 |
0.839 |
0.688 |
|
PRM2 Correlation Coefficient (r) |
0.839 |
1.000 |
0.852 |
|
SEMG1 Correlation Coefficient (r) |
0.688 |
0.852 |
1.000 |
|
Table 5 Correlation studies between the different investigated parameters in the different studied groups
**Statistical significance was set at p≤0.05
PRM1=Protamine-1, PRM2=Protamine-2 and SEMG1 =Semenogelin-1, N= number
A second statistical significant positive correlation existed between count/ml in million and PRM1, PRM2 and SEMG1 levels (r=0.842, r=0.810 and r=0.687 respectively and p<0.000** for all) as shown in Figure 2-4 respectively.
Another statistical significant positive correlation existed between motility and PRM1, PRM2 and SEMG1 levels (r=0.786, r=0.742 and r=0.624 respectively and p<0.000** for all).
Similarly, a statistical significant positive correlation existed between abnormal forms count and PRM1, PRM2 and SEMG1 levels (r=0.489, r=0.502 and r=0.380 respectively and p<0.000** for PRM1 and PRM2 and p<0.007 for SEMG1).
A statistical significant positive correlation was found between the PRM1 levels and both PRM2 and SEMG1 levels (r=0.839 and r=0.688 respectively and p<0.000** for both) as shown in Figures 5,6.
A similar statistical significant positive correlation was found between the PRM2 levels and SEMG1 levels (r=0.852 and p<0.000**) as shown in Figure 7.
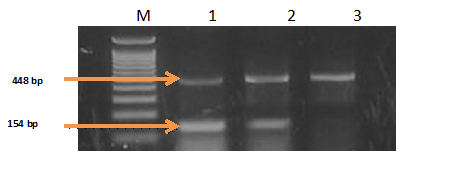
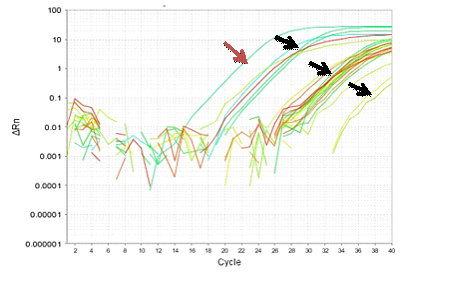
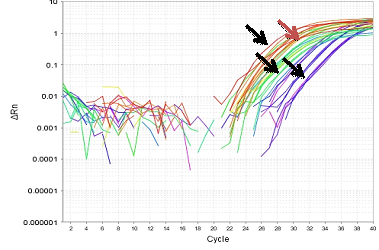
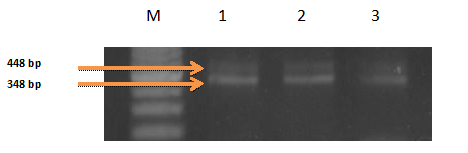
Figure 8-13 showed the PCR results for PRM1, PRM2 and SEMG1 identification in semen contained forensic samples in the three studied groups (normozoospermic, oligozoospermic and azoospermic) as well as their amplification plot curves. PRM1 has got the most clearly visible bands followed by PRM2 and then SEMG1. Also it was clear that in the gel electrophoresis for PRM1 Figure 8, the visibility of the bands decreased with the sperm count with absence of PRM1 band in azoospermic group. It was clear that RT-PCR was more accurate than electrophoresis as it can detect the PRM1 in the azoospermic group Figure 9. As regards PRM2, the band was faint in azoospermic group Figure 10 which indicates that the markers are not only used for identification but also for determining the semen profile of the suspect.
As regards blood samples which were taken from 10 of the normozoospermic studied subjects randomly to test for the presence of the markers of semen, it is noteworthy to mention that none of these markers were detected in the blood, that is to say PRM1, PRM2 and SEMG1 are specific to semen.The presence of certain body fluids in the crime scene can be used as excellent indicators of the sequence of events which occurred. For example, blood stains can indicate some form of physical struggle, assault or murder, and detection of semen or vaginal fluid can indicate the involvement of some form of sexual encounter or assault.14
Recently, the use of a molecular genetics-based approach using messenger RNA (mRNA) profiling has been proposed to supplant conventional methods for body fluid identification.15
Recent advances in forensic genetics have led to the development of several new methods, and the majority of these methods involve the detection of tissue-specific messenger RNA (mRNA) and micro RNA (miRNA) expressions, as well as differential DNA methylation patterns. Especially, recent approaches based on tissue-specific mRNA or miRNA expression have been proved to be useful because of their high tissue specificity to forensically relevant body fluids. With current methodology, DNA profiling can identify an individual from a sample of biological material but it does not reveal what body fluid or tissue source the DNA profile originated from. A multiplex PCR system using messenger RNA (mRNA) has been developed that can identify blood, saliva, semen and menstrual blood in individual stains or in mixtures of body fluids. Messenger RNA transcripts specific to each type of body fluid have been identified and a multiplex reverse transcriptase-polymerase chain reaction (RT-PCR) system developed to identify these body fluids along with three housekeeping genes. This multiplex can detect semen and seminal fluid (semen without spermatozoa present). Furthermore, they have targeted the co-isolation of RNA and DNA from the same sample and, with the RT-PCR multiplex have been targeted, so that the type of body fluid present can be determined as well as generate a DNA profile (s) from the same stain.16 Real-time RT (reverse transcription)-polymerase chain reaction (qRT-PCR) has become the standard for the detection and quantification of RNA targets and is firmly established as a main stream research technology.17
The results of the present study concluded that the three RNA markers (protamine 1, protamine 2 and semenogelin 1) were only detected in semen and not in blood so; these markers can be specifically used for semen identification at the crime scene.
These results are concomitant with those of Juusola et al.6 who proposed a multiplex RT-PCR method for the identification of body fluids that are commonly encountered in forensic casework. Using eight selected body fluid-specific genes, i.e., spectrin (SPTB) and porphobilinogen deaminase (PBGD) for blood, statherin (STATH) and histatin 3 (HTN3) for saliva, protamine 1 (PRM1) and protamine 2 (PRM2) for semen, and human beta-defensin1 (HBD-1) and mucin 4 (MUC4) for vaginal secretions, they were able to detect each of the four body fluids as single or mixed stain.
Similar results were previously obtained by Sakurada et al.7 who have demonstrated the feasibility of identifying body fluids by using a real-time RT-PCR assay where Statherin (STATH) and histatin (HTN3) were the selected genes for saliva and protamine 2 and semenogelin 1 were the selected for semen. This assay showed high specificity to these body fluids thus; both STATH and HTN3 showed specific expression in all saliva samples only. This indicates that these target genes would be effective markers for saliva identification. On the other hand, although both PRM2 and SEMG1 showed specific expression in all semen samples, they also showed a little expression in 2 of the 5 urine samples. This is presumably because urine often becomes contaminated by semen; however, it is certain that PRM2 and SEMG1 have high expression levels in semen.
Similarly, Lindenbergh et al.18 described the development of a single multiplex mRNA-based system for the discrimination of the most common forensic body fluids as well as skin cells. A DNA/RNA co-isolation protocol was established that results in DNA yields equivalent to their standard in-house validated DNA extraction procedure which uses silica-based columns. For semen analysis the markers PRM1 and SEMG1 were selected. They found that PRM1 mRNA was only expressed in fertile men (6 samples), while SEMG1 mRNA was present in semen of both fertile and vasectomised men (2 samples). The assay had good sensitivity as full RNA profiles for blood; semen and saliva were obtained when using 0.05 ml body fluid starting material whereas full DNA profiles were obtained with 0.1 ml.
Moreover, the results of the current study agreed with the results of Nussbaumer et al.10 who reported in their multiplex qRT-PCR assays, which included hemoglobin A (HBA) for blood, kallikrein (KLK) for semen and mucin 4 (MUC4) for vaginal fluid or saliva that, all semen samples showed a high expression of KLK mRNA, while none of other body fluids showed any expression of KLK.
In the study of Conti et al.19 three candidate blood assays; hemoglobin beta (HBB), antigen gamma polypeptide (CD3G), spectrin beta erythrocytic (SPTB) and three semen assays kallikrein 3 (PSA), protamine 2 (PRM2), trans glutaminase 4 (TGM4), demonstrated how minute volumes of blood could be detected using mRNA profiling. Amplification with the HBB probe/primers occurred with as little as 0.0001μl of blood. Although the seminal fluid genes were not as sensitive as the HBB assay, amplification using the PSA and PRM2 TaqMan® sets occurred with 0.01μl of semen. Their experiments demonstrated that TGM4, PSA and PRM2 are robust candidates for the detection of semen or sperm.
Takasakaa et al.20 used the new genome profiling (GP) method in their study to identify sperm and vaginal fluid from RNA extracted from body fluids. Their results suggested that the different profiles of semen and vaginal fluid generated by the GP method could be used to identify the presence of these body fluids in a mixed sample taken from the scene of a sex crime.
The present study concluded that in case we have to select only one marker to search for in the seminal stain, this marker will be protamine 1 since it showed the highest levels of expression among the investigated markers and it also showed the best visible band when gel electrophoresis was done. PRM1 was the best investigated parameter in discrimination between azoospermic, oligospermic and normospermic samples.
The findings of the current study agreed with the results of Lindenbergh et al.18 who assessed the specificity of semen markers (PRM1 and SEMG1) and found that both markers exhibited a high level of specificity with generally lower expression levels for SEMG1 than for PRM1.
Similarly, Conti et al.19 and Sakurada et al.7 detected high specificity of PRM2 and SEMG1 when the sensitivity and specificity of target genes were examined in body fluid stains. The transcripts PRM1, together with PRM2, codes for protamines which are responsible for packaging the sperm chromatin tightly and are important for protecting the paternal genome.21 The protamine 1 and 2 have been studied extensively in relation to male infertility, and an altered protamine1:protamine2 ratio at the protein level has been associated with reduced male fertility.22
In the current study, the results revealed that the markers of semen were not only used for identification of the suspect but also for the differentiation of different semen analysis since positive correlations were found between the markers and sperm count, motility and abnormal forms. A positive correlation detected between count/ml in million and PRM1, PRM2 and SEMG1 levels (r=0.842, r=0.810 and r=0.687 respectively and p<0.000** for all). Another positive correlation existed between motility and PRM1, PRM2 and SEMG1 levels (r=0.786, r=0.742 and r=0.624 respectively and p<0.000** for all). Similarly a positive correlation existed between abnormal forms and PRM1, PRM2 and SEMG1 levels (r=0.489, r=0.502 and r=0.380 respectively and p<0.000** for PRM1 and PRM2 and p<0.007* for SEMG1).
Furthermore, the results of the study showed that the expression of PRM1 and PRM2 was high in cases of normozoospermic men, lower in cases of oligozoospermic and much lower in cases of azoospermic men. The expression of SEMG1 was generally low in most of the cases showing signals in cases of azoospermia, this is because SEMG1 is not a sperm marker but it is a seminal plasma marker. Expression of PRM1 and PRM2 in azoospermic men could be explained on the basis of the study of Marques et al.23 who retrieved testicular sperms from testicular biopsies of azoospermic patients because azoospermia may be due to an ejaculation (mainly due to spinal cord injuries) or due to secondary ejaculatory duct obstruction by infection or primary obstruction azoospermia due to congenital absence of vas deference. It may be secretory azoospermia due to hypospermatogenesis.24
The results of the present study agreed with the results of Haas et al.25 who established RT-PCR protocol for the detection and differentiation of sperm and seminal plasma. Semen specificity of the mRNA markers was successfully confirmed with single-plex PCR. Their data indicated that semen samples down to 0.1ml and up to 20-year-old could be identified with mRNA profiling. Semen samples were taken from 6 normozoospermic and 1 only azoospermic man for analyzing 2 sperm markers (PRM1, PRM2) and 2 novel seminal plasma markers (SEMG1, PSA) concluding that samples from azoospermic men are clearly distinguishable from those of normozoospermic men.
The use of sperm RNA to predict fertility and sperm-quality has been investigated, and several studies have characterized and compared sperm RNAs between fertile and infertile men. An interesting finding is the presence of different transcript levels in sperm of different motility.26-28 Kempisty et al.29 and Jodar et al.30 found a higher content of PRM1 and PRM2 transcripts in spermatozoa of normozoospermic men versus the azoospermic men and suggested that these genes could be a useful marker for predicting male infertility.
Protamine-1 (PRM1), protamine-2 (PRM2) and semenogelin-1 (SEMG1) were only detected in semen samples and were totally absent from blood samples. PRM1 is the most specific and reliable marker used for semen identification. PRM1, PRM2 and SEMG1 are not only used for semen identification at the crime scene but also for determining the semen profile of the suspect.
None.
The author declares no conflict of interest.

©2015 Nader, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.