MOJ
eISSN: 2379-6294


Research Article Volume 5 Issue 1
1Associate Professor from Department of Chemistry, D.A-V, College Kanpur, India
2Assistant Professor from Department of Zoology, D.A-V College Kanpur, India
3Associate Professor from Department of Chemistry, D.A-V, College Kanpur, India
4Department of Biotechnology in Bioinformatics Infrastructure Facility, Forest Research Institute, India
Correspondence: Alka Dubey, Visiting Research Associate, in Bioinformatics Infrastructure Facility (Funded by Dept. of Biotechnology, Govt. of India) of Forest Research Institute, Dehradun, India
Received: January 21, 2019 | Published: February 25, 2019
Citation: Srivastava V, Agrawal SP, Kalra SJS, et al. Modern approaches for drug validation on behalf of computer aided drug designing for Adenocarcinoma. MOJ Toxicol. 2019;5(1):53?60. DOI: 10.15406/mojt.2019.05.00153
In this research our team working on drug validation on behalf of CADD for Adenocarcinoma. We modified some drugs which are already available in market for chemotherapy of Adenocarcinoma. Lung cancer can cause certain changes in the DNA of lung cells. These changes can lead to abnormal cell growth and, sometimes, cancer. In this research we updated well known drug of Adenocarcinoma treatment via computational platforms and we found this drug via protein ligand interaction with favorable statical & structural (Tables Below) computation platforms.
Keywords: Adenocarcinoma, CADD (computer aided drug designing), DNA, drugs
By definition, invasive lung Adenocarcinoma is a malignant epithelial tumor with glandular differentiation, and with either mucin production or pneumocyte marker expression.1 Pulmonary Adenocarcinoma has become the most prevalent histological type of primary lung cancer accounting for almost half of all lung cancers. It is also the most histologically variable and heterogeneous form of lung cancer. This makes it a major focus of research to improve lung cancer patient survival.2 According to the Finnish Cancer Registry, 36% of the histologically confirmed lung cancers in 2007–2012 were reported as Adenocarcinoma, 27% as SCC, and 19% as SCLC Only two decades earlier, SCC was the most common histological type in an epidemiological lung cancer study representing the general population of Northern Finland with a prevalence of 40%, while the prevalence of Adenocarcinoma and SCLC was 26% and 24%, respectively.3 The etiological factors influencing the shift in the relative proportions of pulmonary Adenocarcinoma vs. SCC are complex and not clearly understood.4 In the literature, the emerging predominance of Adenocarcinoma since the 1960s has been strongly related to three smoking-associated factors. First, the change in cigarette manufacturing with the rise of filtered, lower tar- and nicotine containing cigarettes leading to deeper inhaling and a more peripheral distribution of tobacco smoke in the lung.5 This together with the increase in tobacco-specific N-nitrosamines in the manufactured cigarettes has been said to promote a shift from central tumors, including SCC and SCLC, to peripheral tumors, i.e., Adenocarcinoma.6 Second, the risk of SCC and SCLC increases more rapidly with increasing smoking duration than the risk of Adenocarcinoma, causing Adenocarcinoma to appear later.7 Third, the risk of SCC and SCLC decreases more rapidly after smoking cessation than for Adenocarcinoma There is also evidence that non-smoking related factors have influenced the changes in the prevalence of Adenocarcinoma It is estimated that 10–15% of lung cancer deaths are accounted for by factors other than active smoking.8 The improvements in the imaging and detection of peripheral pulmonary nodules as well as changes in the histological classifications 37 of lung tumors and in the pathological techniques may have influenced time trends in the adenocarcino.9 Yet the temporal and geographical patterns and trends observed suggest genuine changes in the prevalence rates Among women, however, Adenocarcinoma rates have always been higher than SCC rates regardless of the smoking status, and the differences have widened over time.10
Cisplatin is similar to the bifunctional alkylating agents. It covalently binds to DNA and disrupts DNA function. After cisplatin enters the cells, the chloride ligands are replaced by water molecules. This reaction results in the formation of positively charged platinum complexes that react with the nucleophilic sites on DNA. These platinum complexes covalently bind to DNA bases using intra-strand and inter-strand cross-links creating cisplatin-DNA adducts thus preventing DNA, RNA and protein synthesis.6 This action is cell cycle phase-nonspecific. Cisplatin also has immunosuppressive, radiosensitizing, and antimicrobial properties. Nephrotoxicity is a major concern when prescribing cisplatin. Renal dysfunction due to cisplatin may manifest as renal insufficiency, hypokalemia and hypomagnesemia. The risk for these adverse effects is related to the dose and interval of cisplatin and may be minimized by adequate hydration.11 Vinorelbine is a semisynthetic vinca alkaloid derived from vinblastine. Vinca alkaloids such as vincristine and vinblastine are originally derived from periwinkle leaves (vinca rosea). Vinorelbine inhibits cell growth by binding to the tubulin of the mitotic microtubules. Like other mitotic inhibitors, vinorelbine also promotes apoptosis in cancer cells. In vitro vinorelbine shows both multidrug and non-multidrug resistance. Mild to moderate peripheral neuropathy (paresthesia, hypesthesia) is the most frequently reported neurologic toxicity and usually reversible on discontinuation of vinorelbine. Cisplatin does not appear to increase the neurotoxic effects of vinorelbine. However, prior treatment with paclitaxel may result in cumulative neurotoxicity.12
The results analysis base on Sequence of Adenocarcinoma antigen ART1 [Homo sapiens] >AAG28523.1 Adenocarcinoma antigen ART1 [Homo sapiens]
MNLQRYWGEIPISSSQTNRSSFDLLPREFRLVEVHDPPLHQPSANKPKPPTMLDIPSEPCSLTIHTIQLI
QHNRRLRNLIATAQAQNQQQTEGVKTEESEPLPSCPGSPPLPDDLLPLDCKNPNAPFQIRHSDPESDFYR
GKGEPVTELSWHSCRQLLYQAVATILAHAGFDCANESVLETLTDVAHEYCLKFTKLLRFAVDREARLGQ
TPFPDVMEQVFHEVGIGSVLSLQKFWQHRIKDYHSYMLQISKQLSEEYERIVNPEKATEDAKPVKIKEEP
VSDITFPVSEELEADLASGDQSLPMGVLGAQSERFPSNLEVEASPQASSAEVNASPLWNLAHVKMEPQES
EEGNVSGHGVLGSDVFEEPMSGMSEAGIPQSPDDSDSSYGSHSTDSLMGSSPVFNQRCKKRMRKI
Expectation Value=0.002, Search Tool = blast, Mask Low Complexity=yes) via BLASTP .this blast mainly use for protein the results of Computer Aided Drug Designing of drug validation to find the potential drug candidate for Adenocarcinoma based on different type potential parameters Homology modeling, also known as comparative modeling of protein, refers to constructing anatomic-resolution model of the “target” protein formats amino acid sequence and an experimental three-dimensional structure of a related homologous protein. In this project homology modeling completed with Geno3D & Phyre it is an automatic web server for protein molecular selected model templates analysis via previous step submission ion Geno 3d server (Figure 1) (Tables 1-4).
Name of template |
Secondary information |
Identity |
pdb4wv4B_0 |
78.4 % |
25.6 |
pdb6mzdC_0 |
98.4 % |
23.6 |
pdb6mzlC_0 |
94.9 % |
23.6 |
Table 1 This table explains about the selected template identity information
Deviation between templates on this chain (Angstrom) |
|||
Name of templates |
4wv4B |
6mzdC |
6mzdC |
4wv4B |
0.00 |
1.15 |
1.15 |
6mzdC |
1.15 |
0.00 |
0.00 |
6mzlC |
1.15 |
0.00 |
0.00 |
Mean deviation : 0.767372 |
|||
Table 2 This table explains structure of statical deviations
Model number |
Models energy (kcal/mol) |
Core |
Allowed |
Generously |
Disallowed |
Model 1 |
8244.74 |
80.2% |
16.0% |
3.7% |
0.0% |
Model 2 |
-3567.42 |
80.2% |
16.0% |
1.2% |
2.5% |
Model 3 |
-3486.13 |
90.1% |
4.9% |
2.5% |
2.5% |
Model 4 |
-3519.33 |
80.2% |
17.3% |
2.5% |
0.0% |
Model 5 |
-3597.54 |
87.7% |
11.1% |
1.2% |
0.0% |
Model 6 |
-3536.62 |
82.7% |
13.6% |
2.5% |
1.2% |
Model 7 |
-3319.28 |
86.4% |
9.9% |
2.5% |
1.2% |
Model 8 |
-2316.14 |
17.3% |
44.4% |
22.2% |
16.0% |
Model 9 |
-3393.46 |
82.7% |
12.3% |
2.5% |
2.5% |
Model 10 |
-3509.73 |
88.9% |
8.6% |
1.2% |
1.2% |
Table 3 This table explains the energy and core of each predicted models
Model validation completed with the help of SAVES server. It is type of online web server for model validation and for analyzing protein structure for validity and assessing how correct they are it’s based on six programs Results: the Model 1 is pass by the SAVES server as per selection of model numbers we analyze the quality factors of model number one & eight for potential selection and analysis (Table 5).

Table 5 This table explains about the quality factor analysis on behalf of CADD paramets for the potential analysis
Visualization
Selected model analysis based on 2D and 3D structure visualization and the model visualization completed with the help of different type of software’s for example: Chimera, Rasmol, and discovery studio computational predicted models via bioinformatics approaches and the method of homology modelling is based on the observations because that’s protein tertiary structure is better conserved than amino acid sequence (Table 6).
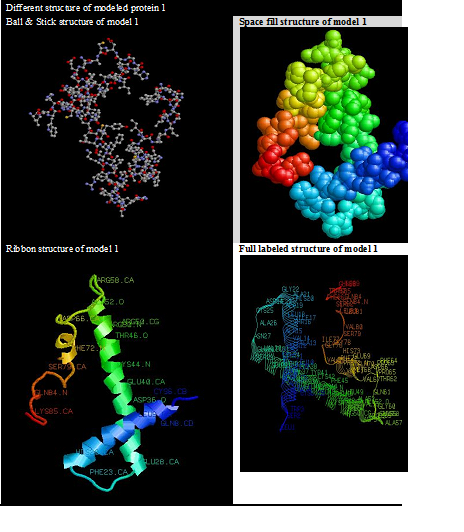
Table 6 This table explains about the different variants of selected models with labeled information
Suitable ligand selection for receptor ligand binding
In this project the ligand selection for potential receptor based on ligand binding site and also available list favorable drugs for Adenocarcinoma Then select the potential ligand as drug candidate and change some confirmation on drug structure computationally then prepare final ligand for docking results analysis (Table 7).

Table 7 This table explains about the different variants of selected ligand molecules with some modification on behalf of CADD with labeled information
Docking
Prediction of the optimal physical configuration and energy between two molecules and the phenomenon which enables the interaction between receptor molecules and the ligand molecule, and mainly (Tables 8-10).
Docking Model Number |
Binding Score |
Area |
ACE |
Transformation |
Docking Model 1 |
6484 |
969.40 |
-488.42 |
-0.34 -0.89 -2.33 -0.66 1.16 14.47 |
Docking Model 2 |
6092 |
858.50 |
-516.24 |
0.87 0.15 -0.84 26.94 -8.85 -2.40 |
Docking Model 3 |
6038 |
770.60 |
-538.41 |
1.91 1.03 -1.12 10.82 -8.62 17.59 |
Docking Model 4 |
5956 |
745.90 |
-386.03 |
-0.91 -0.03 2.24 28.20 -9.65 -2.88 |
Docking Model 5 |
5948 |
791.90 |
-409.23 |
-0.49 0.88 2.02 -2.40 1.67 12.96 |
Docking Model 6 |
5902 |
845.40 |
-375.84 |
0.64 0.74 0.54 -2.93 0.89 13.01 |
Docking Model 7 |
5860 |
739.70 |
-477.23 |
-1.75 -0.95 2.28 10.98 -7.33 15.59 |
Docking Model 8 |
5702 |
810.00 |
-349.13 |
0.31 -0.77 -1.49 -2.45 1.06 11.90 |
Docking Model 9 |
5648 |
888.80 |
-533.52 |
0.45 0.96 0.86 0.75 1.34 15.40 |
Docking Model 10 |
5644 |
769.10 |
-363.72 |
-0.71 -0.63 2.50 24.25 -7.03 -4.95 |
Docking Model 11 |
5618 |
908.10 |
-590.07 |
1.14 -0.53 -0.39 -3.03 0.06 13.77 |
Docking Model 12 |
5616 |
738.90 |
-359.77 |
-1.00 0.10 2.65 1.32 0.60 12.19 |
Docking Model 13 |
5596 |
712.70 |
-348.23 |
0.49 0.71 -0.43 24.10 -5.34 -4.50 |
Docking Model 14 |
5594 |
725.20 |
-529.50 |
-1.42 -0.59 2.31 12.78 -10.56 18.25 |
Docking Model 15 |
5556 |
697.30 |
-399.67 |
-2.18 -0.59 -3.08 22.48 -5.53 9.24 |
Docking Model 16 |
5536 |
709.40 |
-420.11 |
1.67 -0.57 -0.92 9.93 -7.17 19.02 |
Docking Model 17 |
5530 |
738.20 |
-428.25 |
-0.75 -0.61 2.52 27.07 -8.49 -2.49 |
Docking Model 18 |
5526 |
758.30 |
-451.96 |
-1.02 -0.61 2.80 27.81 -12.64 -4.91 |
Docking Model 19 |
5500 |
742.50 |
-431.86 |
1.65 0.13 -0.88 8.80 -6.00 14.69 |
Docking Model 20 |
5478 |
740.40 |
-430.40 |
-0.92 0.54 2.35 -1.36 3.82 15.58 |
Table 9 This table explains the results of docking with Adenocarcinoma with modified vinorelbine. Total 20 models predicted with statical parameters but we selected model numbers 1 out of 20 because good bing score and area coverage of the molecule
Docking Model Number |
Binding Score |
Area |
ACE |
Transformation |
Docking Model 1 |
1638 |
178.20 |
-44.46 |
0.27 0.71 0.16 26.54 -10.96 -0.94 |
Docking Model 2 |
1504 |
157.40 |
-30.81 |
-3.02 -0.51 -2.06 -4.00 -6.29 20.85 |
Docking Model 3 |
1438 |
160.90 |
-13.19 |
-2.64 0.83 1.25 -6.84 -0.05 11.30 |
Docking Model 4 |
1436 |
163.50 |
-36.76 |
1.04 0.58 0.36 23.04 -17.00 0.24 |
Docking Model 5 |
1420 |
159.70 |
-29.52 |
-2.94 -1.02 -1.67 0.09 -1.10 13.21 |
Docking Model 6 |
1402 |
149.30 |
-34.38 |
-0.86 -0.01 -2.21 -9.17 1.49 14.51 |
Docking Model 7 |
1396 |
144.80 |
-19.11 |
-2.75 -0.45 -2.61 15.76 -5.16 -9.38 |
Docking Model 8 |
1374 |
151.90 |
-40.61 |
-0.16 0.14 1.92 15.39 -7.33 4.17 |
Docking Model 9 |
1362 |
145.00 |
-36.10 |
-0.90 -0.54 -1.02 5.62 -3.16 22.31 |
Docking Model 10 |
1360 |
156.70 |
-30.83 |
1.33 0.77 2.24 10.56 -10.98 14.40 |
Docking Model 11 |
1354 |
142.30 |
-36.61 |
-0.60 0.33 2.13 18.39 -12.55 4.51 |
Docking Model 12 |
1354 |
143.70 |
-28.74 |
-1.21 -1.11 0.95 14.12 -16.01 19.93 |
Docking Model 13 |
1350 |
148.30 |
-20.09 |
-0.45 -0.12 2.52 -2.01 -3.81 23.86 |
Docking Model 14 |
1330 |
136.30 |
-20.51 |
2.00 1.13 -0.52 7.50 -15.02 5.07 |
Docking Model 15 |
1324 |
145.00 |
-30.90 |
-1.54 -0.51 2.34 -2.05 -13.74 13.49 |
Docking Model 16 |
1324 |
147.10 |
-27.18 |
1.23 1.02 -2.68 24.98 -5.42 9.50 |
Docking Model 17 |
1322 |
142.20 |
-4.96 |
-2.49 -1.45 0.53 8.26 -6.39 -4.67 |
Docking Model 18 |
1318 |
137.30 |
-24.31 |
-1.36 1.12 1.87 8.50 -5.70 19.27 |
Docking Model 19 |
1316 |
136.00 |
-18.42 |
1.16 -0.25 2.57 1.00 -16.18 12.70 |
Docking Model 20 |
1308 |
142.40 |
-25.04 |
2.75 0.36 0.62 4.80 2.11 17.80 |
Table 10 This table explains the results of docking with Adenocarcinoma with modified cisplatin Total 20 models predicted with statical parameters but we selected model numbers 1 out of 20 because good bing score and area coverage of the molecule
Finally in this study we modified cisplatin & vinorelbine chemical structures via online available tools on behalf of CADD parameters and 3Dstructure of Adenocarcinoma protein (homo sapience) which is predicted through the Insilico approaches and homology modeling and the docking of Modified cisplatin & vinorelbine with other selected various ligand and determined the interaction between protein and ligand that’s bind on active site of the Modified cisplatin & vinorelbine, although docking process. It is very complicated because its depends on various parameters the main resultant obtained by different type of docking tools and docking completed with the help of HEX,PATHADOCK for identify the suitable Modified cisplatin & vinorelbine and other different ligands which are docked with the Adenocarcinoma protein for inhibit the growth of unnatural cell development . Only 2 numbers of ligand given the minimum energy out of other selected 10 ligands. Modified cisplatin & vinorelbine is playing an important role as inhibitor for treatment of Adenocarcinoma with minimum binding energy and its work as a potential Inhibitor for unnatural cell development in lungs that causes Adenocarcinoma as per the Table number 9 & 10 parameters. Perhaps the ultimate solution is to develop a potential drug candidate against this devastating unnatural cell development in lungs that causes Adenocarcinoma.20
It is my proud privilege to express deepest sense of reverence and heart full thanks DBT, for giving me this platform I am extremely great full to Er. Neelesh Yadav from BIFC–FRI, Dehradun for giving me valuable guidance related to advance computer technology
Author declare that there is no conflict of interest.

©2019 Srivastava, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.