MOJ
eISSN: 2374-6939


Research Article Volume 3 Issue 5
Victor dupouy hospital, France
Correspondence: Farshid Moshiri, Victor dupouy hospital, 69, rue lieutenant colonnel prud'don 95100 argenteuil, Paris, Tel 33613046420
Received: December 05, 2014 | Published: December 1, 2015
Citation: Moshiri E (2015) Role of High Mobility Group Box 1 in Bone Formation. MOJ Orthop Rheumatol 3(5): 00112. DOI: 10.15406/mojor.2015.03.00112
Endochondral ossification is a multistep process, which involves spatiotemporally strictly regulated morphogenetic and differentiation steps. Chondrogenesis starts with the condensation and commitment of mesenchymal precursors and their differentiation to prechondrocytes in a shape prefiguring the future bones. Chicken embryo chondroblast (CEC), fibroblast (CEF) and high-density mesenchyme (HDM) cultures undergoing chondrogenesis in vitro were prepared. CEC cultures were obtained from sterna of day 14.5 embryos using 0.1 % collagenase treatment. CEF cultures were prepared from 8-10 day embryos by trypsin treatment. HDM cultures were made from the limb buds of stage 23-24 chicken embryos.
Keywords: Chondrogenesis, HMG proteins, Non-histone chromatin components, Enhancers, silencers
CEC, Chondroblast; CEF, Fibroblast; HDM, High-Density Mesenchyme; PTHrP Parathyroid Hormone-Related Protein; Ihh, Indian Hedgehog; EMSA, Electrophoretic Mobility Shift Assay
Cartilage and chondrogenesis
Endochondral bone formation: Endochondral ossification is a multistep process, which involves spatiotemporally strictly regulated morphogenetic and differentiation steps.1,2 It leads to the formation of most of the skeletal elements during vertebrate embryonic development and it takes place also in the growth plates, where it is responsible for longitudinal bone growth till adulthood. During endochondral ossification mesenchymal precursors differentiate into bone tissue through replacement of a cartilaginous intermediate.
Chondrogenesis: starts with the condensation and commitment of mesenchymal precursors and their differentiation to prechondrocytes in a shape prefiguring the future bones (Figure 1). The prechondrocytes subsequently differentiate into early chondroblasts, which proliferate, form columns and deposit an extensive cartilaginous extracellular matrix (ECM) serving as template of the future bones. Then, starting from the middle of the cartilaginous primordia, proliferative chondroblasts exit from the cell cycle and successively undergo prehypertrophic and hypertrophic development.3,4 Chondrogenesis is directed by three master transcription factors, Sox9, L-Sox5 and Sox6, called the Sox trio (Figure 1).5 Numerous autocrine, paracrine and endocrine factors determine the shape and size of skeletal elements often acting by modulating the expression or the activity of the Sox trio. Maintenance of proliferation in the columnar zone and bone growth depends on positive and negative feedback loop formed between Ihh (Indian hedgehog) and PTHrP (parathyroid hormone-related protein) and affected by growth factors (FGF, BMP), hormones and various other signaling pathways.2,3,6 Stimulation of chondrocyte proliferation by mechanical load is mediated by Ihh via BMP signaling.7 On the other hand, induction of Ihh expression in the prehypertrophic zone, hypertrophic differentiation and ossification is regulated by Runx2, the master factor of the osteoblast lineage.6 In addition, Hmgb1 secreted by hypertrophic growth plate chondrocytes is needed for cell invasion to function as a chemoattractant for osteoclasts, osteoblasts and endothelial cells.8,9

Figure 1 Chondrogenesis. Stages of chondrocyte differentiation are shown indicating the up- and down regulation of important marker genes by green and yellow arrows, respectively. Role of Sox transcription factors in the regulation of differentiation steps is also indicated.
Homozygous mice for cmd die at birth and suffer from disproportionate dwarfism, clef palate, short snout and protruding tongue. Targeted inactivation of Crtl1 in transgenic mice revealed the essential role of the protein in cartilage development. Most homozygotes died shortly after birth due to respiratory failure and the few survivors developed progressive drawfism, craniofacial abnormalities and lordosis of the cervical spine. They showed characteristics of spondyloepiphyseal dysplasias, such as small epiphysis and flattened vertebrae.10
Mutations in other cartilage proteins involved in the ECM assembly or disruption of their genes have less dramatic effects in animal models. For instance, mice deficient in matrilin family members are viable under laboratory conditions.11 Matn1 up regulation was implicated in vertebral fusion of Atlantic salmon.12 A linkage of MATN1 was reported to osteoarthritis in the Dutch population.13 Various polymorphism of MATN1 was found to predispose to idiopathic scoliosis in Asian and Turkish population.14 MATN1 polymorphism was also linked to mandibular prognatism in human.15 MATN1 was clearly associated to relapsing polychondritis, a rare autoimmune disease. Circulating MATN1 antibodies were found to function as auto antigen in these patients, and immunization of mouse or rat with MATN1 caused similar symptoms in an animal model of this human disease.16,17
The non-histone chromatin components function as architectural factors in the organization and fluidization of the chromatin by appropriately bending and plasticizing DNA. For instance, the high mobility group (HMG) proteins belong to three families based on their DNA-binding domains. Hmga proteins interact with AT-rich sequences in the minor groove of the DNA by their AT-hook HMGA domain. Hmgb proteins bind DNA without sequence specificity in the minor groove by their L-shaped HMG box (HMGB) domains. Hmgn proteins directly interact with nucleosomes via their nucleosome-like HMGN domain. HMGBs better bend the DNA than transcription factors that bind DNA in the major groove.
Proximal and distal cis-control elements are critical players of tissue-specific gene regulation. The proximal DNA elements can function in one orientation and only within a few hundred bp from the TATA box. They usually have binding sites for some ubiquitous and tissue-specific factors. By contrast, enhancers and silencers highly activate or repress transcription, respectively, over large distances and independent of orientation. They usually carry an array of recognition motifs for a unique set of transcription factors, which bind in a cooperative manner and interact with each other and other factors and GTFs bound to proximal DNA elements and the core promoter. Thus, enhancers perform a crucial function in the formation of stereospecific nucleoprotein complexes, the enhance osomes, which modulate transcription via very complex protein-protein and protein-DNA interactions. Tissue-specific gene regulation is based on a combinatorial interaction between cis-acting elements and trans-acting factors. Apart from classical transcription factors, recent data underline the important role of SOX proteins and canonical HMGB proteins in the regulation of gene expression during development and disease.
L-Sox5 and Sox6 are expressed in early proliferative chondroblasts and they are required for the differentiation of early chondroblasts to late proliferative or columnar chondroblasts in the growth plate. L-Sox5 and Sox6 are also needed for the activation of some of the cartilage ECM genes (Comp, Matn1) and for the high activity of others (Col2al, Acan, etc.) L-Sox5 is a longer product of the Sox5 gene. It is chondrocyte-specific splice variant and highly similar to Sox6. Sox5-null and Sox6-null mice are born with minor cartilage defects, whereas the double-null mice develop a severe, generalized chondrodysplasia and die around embryonic day 16.5.18 Respectively, these mice have impairment of chondroblast proliferation and expression of cartilage matrix genes. Interestingly, L-Sox5 and Sox6 are required to turn on Matn1, as Matn1mRNAwas not detected in Sox5-/-; Sox6-/- mice (Figure 1).18
Aim of this study is that among the cartilage protein genes, Matn1 has a unique expression pattern, restricted to certain chondrocyte developmental stages and distinct zones of the growth plate. This suggests that regulation of the Matn1 may involve similar as well as different molecular mechanism as compared to other cartilage protein genes, e.g. Col2a1. To get insight into the common and distinct molecular mechanisms controlling cartilage-specific gene expression, we aimed to identify DNA elements and uncover the role of transcription factors in the transcriptional regulation mechanism of Matn1.
Cell culture
Chicken embryo chondroblast (CEC), fibroblast (CEF) and high-density mesenchyme (HDM) cultures undergoing chondrogenesis in vitro were prepared as described. CEC cultures were obtained from sterna of day 14.5 embryos using 0.1 % collagenase treatment. CEF cultures were prepared from 8-10 day embryos by trypsin treatment. HDM cultures were made from the limb buds of stage 23-24 chicken embryos. Low-density mesenchyme (LDM) cultures were made similarly as HDM cultures, but instead of 5x106 cells, only 1x106 cells were plated in 35-mm plates in F12/DMEM 1:1 (HyClone Laboratories) supplemented with 10% FBS (Sigma and GIBCO Laboratories). COS-7 cells were cultured under standard conditions. HDM cultures consisting of early proliferative (stage Ia) chondroblasts and CEC cultures rich in late proliferative (stage Ib) chondroblasts represented the low and high Matn1-expressing cell types, respectively.19,20 LDM, CEF, COS-7 cultures were used as Matn1-nonexpressing controls. The C-28I/2 immortalized human costal chondrocyte, the SW1353 human chondrosarcoma (ATCC HTB-94) and the RCS (rat chondrosarcoma) cell lines were cultured in DMEM supplemented with 10% FCS (GIBCO).
Quantitative real-time PCR (QRT-PCR)
Total RNA was isolated from CEC, CEF or HDM cultures at subsequent days of chondrogenesis using the RNA isolation kit (Macherey-Nagel) according to the manufacturer’s instructions. The quantity of isolated RNA was measured by spectrophotometry (NanoDrop). QRT-PCR was performed on a Rotor Gene 3000 instrument (Corbett Research) with genes pecific primers and SybrGreen protocol to monitor gene expression changes. The primers were designed by the Roche online primers design tool (https://www.roche-appliedscience.com/sis/rtpcr/upl). Briefly, 2 μg of DNase-treated total RNA from each sample was reverse transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems) according to the manufacturer’s instructions. Reactions were done with Fast Start SYBR Green Master mix (Roche Applied Science) at a final primer concentration of 250 nM as follows: 15 min at 95oC, 45 cycles of 95oC for 15 s, 60oC for 25 s and 72oC for 25s. The quality of the reaction was checked by melting temperature analysis after each reaction. The quality of primers was verified by MS analysis provided by Bioneer (Daejeon). Each individual Cτ values were normalized to the average Cτ values of three internal control genes (GAPDH, 18S rRNA, and 28S rRNA). The final relative gene expression ratios were calculated as either 2−ΔCτ values (compared to the internal control genes) or 2−ΔΔCτ values (comparison of the normalized ratios) as indicated in the figure legend.
Plasmid constructions
∆IneM3- and∆SI2dm-AC8Luc were made by the PCR-based Quick ChangeTM Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions using AC8Luc as template and oligonucleotides IneM1, IneM2, IneM3 and P1-2d carrying mutation in the Nfi binding site of SI, respectively. Double mutants ∆Pe1M1-∆IneM2-, ∆Pe1M1-∆SI2dm- and ∆Pe1M4-∆SI2dm- AC8Lucwere constructed similarly using ∆Pe1M1- and ∆Pe1M4FO15Luc DNAs as templates. ∆Dpe1ABC- and ∆Dpe1BC- were made by deleting sequences between positions −1879/−1791 and −1848/−1791, respectively, from the long Matn1 promoter. Mutantderivatives of8xECol2a1-FO15Luc were made by replacing the wild-type short promoter of 8xECol2a1-FO15Luc with the corresponding mutant FO15Luc fragment. Structures and sequences of all constructs were verified. Luciferase reporters harboring multiple copies of the Dpe1 element upstream of the Matn1 short promoter were made by inserting four copies of the PCR-amplified Dpe1 fragment into FO15Luc. 4×Dpe1(−)FO15Luc was generated by inserting blunted four copies of Dpe1 into FO15Luc in reverse orientation. PCLuc and 4×Dpe1(+)PCLuc were generated by replacing the Matn1 short promoter of FO15Luc and 4×Dpe1(+)FO15Luc, respectively, with the Col2a1 short promoter fragment between positions −309/+118. Structures and sequences of all constructs were verified by restriction mapping and sequencing.
Transient expression assays
CEC and CEF cultures were transfected with 2 µg reporters, while HDM, LDM, and COS-7 cultures were transfected with 5µg reporters using the Ca-phosphate coprecipitation method as described previously. Cotransfection with pRL-TK (Promega) served as an internal control to correct for transfection efficiency, but parallel plates were also transfected with FO15Luc. Firefly and renilla luciferase activities were measured in a Luminoscan Ascent (Thermo Lab system 2.6) using the respective Luciferase Assay Systems (Promega) according to the supplier’s instructions 72 h (HDM and LDM) or 48 h (other cells) posttransfection. Relative luciferase activities were expressed in fold as compared to values of FO15Luc taken as 1, unless noted otherwise.
Electrophoretic mobility shift assasy (EMSA)
Nucleotide sequences of Ine and Pe1 described previously, and that of Dpe1 is presented later in Figure 2A. Double-stranded oligonucleotides were synthesized for the Dpe1 element comprising positions −1879/−1791: 5'-GAG TCC AGT GTT TTC GTT TTT GGA GGC CCG GGG AA-3’ (Dpe1A), 5'-GGA AAA ATT ATG TTT CAT ATA TTA AAA ATA AAC A-3’ (Dpe1B), 5'-AAA TAA ACA CTA CTT TTA CAG AGG TAT AAA TGC-3’ (Dpe1C). Coding region of Hmgb1 was inserted in frame into pGEX expression vector. GST-tagged L-Sox5, SOX9 and Hmgb1 were expressed and purified, and crude cell extracts were made as described. 20-30 fmol end-labeled DNA probes were incubated either with 0.6-3.2μg purified GSTfused Hmgb1, SOX9, L-Sox5 or 3 μg crude CEC or CEF cell extracts in the presence of 100-500 ng poly(dG-dC)·(dG-dC) and separated on pre-run 5% or 6.6% PAGE.

Figure 2 Dose-dependent activation of the Matn1 promoter and Dpe1 fused to homologus or heterologous short promoters by the Sox trio. (A-D) AC8Luc was cotransfected with Sox expression plasmids in various mesenchymal cultures as indicated (A-B) and COS-7 cells (C-D). Combined forced expression assay and Western analysis with anti-FLAG antibody (C). Forced expression assays were performed with constant amount of FLAG-tagged expression plasmid for SOX9 and increasing amount of expression plasmids for L-Sox5 and Sox6. (E) Map of the reporters driven by four copies of Dpe1 fused to the homologous Matn1 or the heterologous Col2a1short promoters in direct or reverse orientations as indicated. (F-G) Dose-dependent synergistic activation of these reporters by forced expression of the Sox trio. Luciferase activities are presented as fold values relative to that for AC8Luc (A-D), FO15Luc (F) and PCLuc (G). *p<0.05, **p<0.01, ***p<0.001 compared with reporter cotransfected with vectors (A-D) or vs. vector-cotransfected 4xDpe1-reporters (F-G); #p<0.05, ##p<0.01, ###p<0.001 compared with SOX9 cotransfected reporters (A-D) or as indicated (C,F and G).
Immuno fluorescence
Acetone-fixed 10-µm cryosections were used for immunofluorescence. Nonspecific binding of the antibodies was blocked with 10% normal goat serum. The specimens were incubated at 4oC overnight with the following primary antibodies in combinations: rabbit affinity purified antisera specific for Matn1 (1:200 dilution) and SOX9 (Abcam, ab3697, 1:50) and mouse monoclonal antibody for HMGB1 (MBL, M137-3, 1:200). The appropriate secondary antibodies were applied at room temperature for 1 h in the dark: Alexa 488-labeled anti-rabbit IgG antiserum (Molecular Probes, 1:400), Cy3-conjugated anti-mouse IgG antibodies (Jackson Immunoresearch, 1:400). Nuclei were stained with 1 µg/ml Hoechst in PBS for 5 min. The specimens were mounted with fluorescent mounting medium (Dako), viewed with a Nikon Eclipse E600 microscope equipped with epifluorescence and Pan flour objectives, and photographed with Nikon digital camera D5000. After immunofluorescence, the coverslips were removed and the sections were restained with hematoxylin and eosin. The images were processed using SPOT software (version 4.0.9 for Windows; Diagnostic Instruments) and figures were made with Adobe Photoshop 8.0 and CorelDraw X4 soft wares.
Forced expression assays combined with western analysis and QRT-PCR
To estimate the relative expression levels of Sox and Nfi proteins, we used pcDNA5’UTFLAG-SOX9 (pFSOX9)21 and we made pFNfib by inserting the NotI (blunted)-NheI fragment of Nfi expression plasmids22 into the EcoRI (blunted)-XbaI sites of pcDNA5’UTFLAG. COS-7 cells were cotransfected as described above with AC8Luc, 1 µg pFSOX9 and increasing amounts of pFSox5 and pFSox6. In other experiments increasing amounts of pFNfib, effector plasmid was used with 1 µg pFSOX9. In addition, we made effector plasmid pFHmgb1 by inserting the Hmgb1 coding region into pcDNA5’-UT-2FLAG. To estimate the relative expression levels of Sox and Hmgb1 proteins, we cotransfected COS-7 cells with 10 µg AC8Luc, 1 µg each of pFSOX9, pFSox5 and pFSox6, and increasing amounts of pFHmgb1. The transfected cells were lysed in 100 µl buffer containing 14 mM HEPES (pH 7.9), 1.5 mM MgCl2, 6 mM KCl, 0.44 mM NaCl, 0.08 mM EDTA, 3.5 mM DTT, 0.5 mM PMSF, 10% glycerol and protease inhibitor cocktail (Sigma-Aldrich P2714).
To test the induction of the endogenous Matn1 in forced expression assays, we cotransfected COS-7 cells with 50 ng pFSOX9, 75 ng pFSox5, and 75 ng pFSox6 without and with 800 ng pFHmgb1 using 2µl TurboFect (ThermoScientific, R0531). Transfection mixtures were adjusted with empty vectors to the same amount of total DNA. Transfections were made in duplicates and repeated 3 times. RNA was isolated from the cells and the Matn1 mRNA level was determined by QRT-PCR using the SybrGreen protocol and gene-specific primer pairs (Suppl. Table S1). Cτ values were normalized to that of Gapdh. Data are presented as mean ± SEM.
Hmgb1 silencing
Silencing experiments were performed in chondrogenic cell lines C-28/I2, SW1353, and RCS with siRNAs purchased from Bioneer Corporation (Daejeon, Republic of Korea) for human HMGB1: 5’-caggaggaauacugaacau-3’; for rat Hmgb1: 5’-cugucaacuucucagaguu-3’; for human GAPDH: 5’-gugugaaccaugagaagua-3’, and for negative control si RNA: 5’- ccuacgccaccaauuucgu-3’. 1.2-2.0 x 105 cells were plated in 6-well plates and transfected with 100-400 p moles of si RNA duplexes 24 h after plating using X-tremeGENE siRNA Transfection Reagent (Roche Applied Science) as suggested by the supplier. Cultures were harvested 30 h (RCS) or 42 h (C-28/I2, SW1353) after transfection. RNA was isolated from the cells and marker gene expression levels were determined by QRT-PCR using the Sybr Green protocol and gene specific primer pairs (Suppl. Table S1). Gene expression levels were normalized to the invariant Rps18 mRNA levels. Data are presented as mean ± SEM from three independent experiments.
Accumulation of Nfi and Sox mRNAs during in vitro chondrogenesis
To address the potential contribution of Sox and Nfi factors and Hmgb1, we compared the kinetic changes in the expressionof Matn1, Sox trio, Nfiand other marker genes in various chicken primary cultures by QRT-PCR during in vitro chondrogenesis in HDM culture (Figure 3). This culture faithfully mimics the early steps of chondrogenesis as it differentiates to early proliferative chondroblasts characterized by elevated Col6a1 expression (Figure 3A). CEC culture, expressing the genes for Sox trio and cartilage proteins at high levels (Figure 3A-3C), represented a later stage. CEF culture served as a negative control. In CEF, the steady state mRNA levels for Matn1 and the Sox trio were very low, while those for Hmgb1and Nfi except Nfic were elevated relative to their levels in committed mesenchyme (HDM day 0) (Figure 3B-3E).
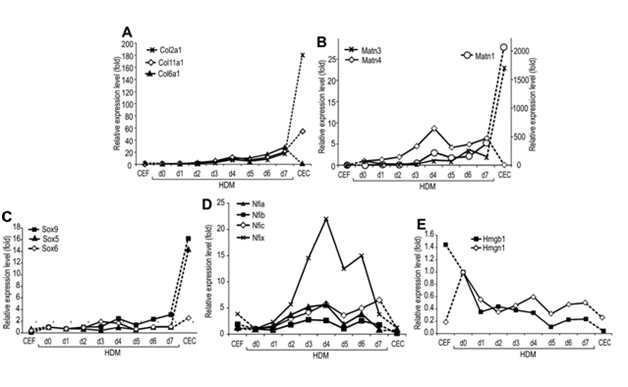
Figure 3 Comparison of the marker gene expression by QRT - PCR during in vitro chondrogenesis (A Marker mRNA levels were determined by QRT-PCR in HDM culture undergoing chondrogenesis in vitro and compared to mRNA levels of non-expressing CEF and high Matn1-expressing CEC cultures. Cτ values were normalized to the average Cτ values of three internal control genes. Relative expression levels are presented as fold values relative to the HDM day 0 values.
Unlike the slow continuous accumulation of Col2a1mRNA during early stages of differentiation in HDM culture, activation of Matn1 was first detected in HDM culture at day 4 (Figure 4A & 4B). Sox9 and Col2a1 mRNAs accumulated with similar kinetics, but the low levels of L-Sox5 and Sox6 mRNAs increased sharply only in CEC culture, except for a small, transient boost of Sox6 mRNA at days 3 and 4 in HDM culture, just preceding the first peak in the Matn1 mRNA level (Figure 4B & 4C). Interestingly, Matn3 and Matn4, although also expressed in cartilage, exhibited smaller increase in their relative expression level than Matn1 (Suppl. Table S2). Matn4level, however, peaked in HDM culture suggesting a function in early stage of chondrogenesis (Figure 4B). Remarkably, from a very low expression level compared to the internal control genes measured in committed mesenchyme (Suppl. Table S2), Matn1expression showed the highest relative increase (2057-fold) in CEC culture, in contrast to the lower increase in the level of Col2a1 (181-fold) and other cartilage ECM genes (<80-fold, compared to HDM day 0) (Figure 4A & 4B). The relative Nfi mRNA levels also increased transiently 2.6 to 22-fold with two peaks at day 4 and days 6-7 in HDM culture, followed by a sharp decline in CEC culture close to 1 (Nfia and Nfix) or below 1 (Nfib and Nfic). In contrast, Hmgb1 mRNA level declined gradually in HDM culture, exhibiting the lowest level in CEC culture. Thus, CEC culture, rich in late proliferative chondroblasts, is characterized by high Matn1 and Sox trio, but low Nfi and Hmgb1 mRNA levels. Day 4 HDM culture, consisting of early proliferative chondroblasts, however, exhibits high Nfi, but lower Matn1, Sox9 and Sox6 mRNA levels and very low L-Sox5 mRNA expression. Sox6 and Nfi mRNA levels peaked in HDM culture at the time of Matn1activation, suggesting a function in Matn1regulation.
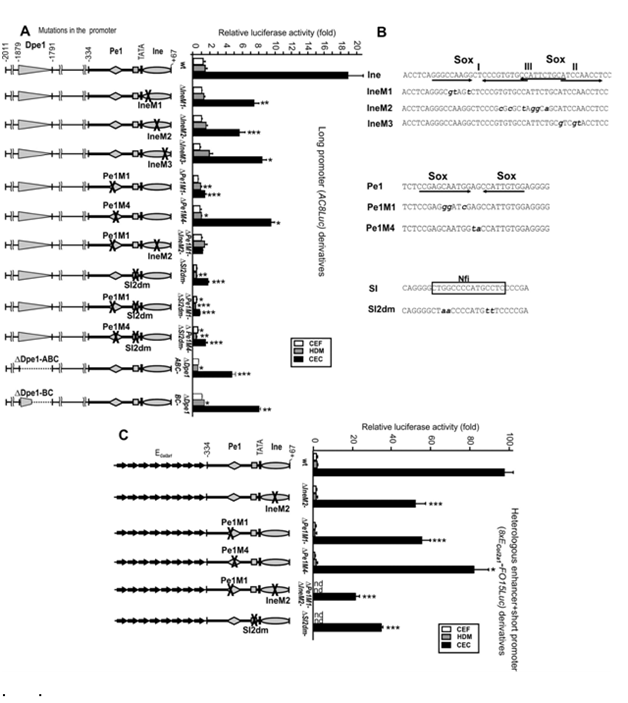
Figure 4 Effect of Ine, Pe1 and SI mutations and Dpe1 deletion on the long promoter activity in mesenchymal cultures. Transient expression activities of wild-type and mutant versions of reporters AC8Luc
Figure 4A 8xECol2a1-FO15Luc (C) driven by long Matn1promoter or multiple ECol2a1 fused to the short promoter, respectively. Schematic to the left indicate single or double mutations introduced into the promoter
Figure 4B Sequences of wild-type or mutant version of DNA elements. Sox- and Nfi-binding sites are indicated (A and
Figure 4C Luciferase activities of wild-type (wt) and mutant reporters in the low-, high-, and non expressing HDM, CEC, and CEF cultures, respectively, are presented in fold relative to that of FO15Luc. *p<0.05, **p<0.01, ***p<0.001 mutants vs. wild-type reporters.
Comparative binding of Sox transcription factors to the conserved Matn1 promoter elements in vitro
This study hypothesized that Sox factor-mediated interactions may play important role in the chondroblast developmental stage-dependent regulation of Matn1. To test this hypothesis, first we compared the binding of purified chondrogenic Sox factors to Dpe1 and the short promoter elements Pe1 and Ine (Figure 4).
We found that the chicken Dpe1 element harbors three putative sites with tandem and inverted paired motifs, which share 5/7 or 6/7 nucleotide identity with the Sox consensus sequence (Figure 4A). These motifs also show 5/10 to 8/10 nucleotide identity with AGAACAATGG, the preferred Sox9-binding site. We performed EMSA experiments to demonstrate that the element carries at least three sites which can interact with purified Sox proteins in vitro. Each of the three subfragments of Dpe1 was clearly recognized by GST-fused SOX9 and L-Sox5, but with inverse binding efficiency (Figure 4B & 4C). SOX9 exhibited the strongest binding to Dpe1C forming two complexes and weaker binding to Dpe1A and Dpe1B forming one and three diffuse complexes, respectively (Figure 4B). On the other hand, L-Sox5 most preferably recognized Dpe1B, followed by A and C (Figure 4C). We found that SOX9 bound the Matn1 control elements with highly variable efficiency. Whereas it showed more potent complex formation in vitro with each of the Dpe1 Sox sites, than with those of Ine, SOX9 bound Pe1 even more powerfully (≥5-fold) than Dpe1C (Figure 4B).
We studied that effect of SI mutation on the long promoter activity. The −2011/−334 sequence enhanced the short promoter activity ~19-fold in AC8Luc in CECs, but hardly did so in low- or non-expressing cultures (Figure 4A). Sox site mutations IneM1, IneM2 and IneM3 cut the long promoter activity to half or more in CECs. The effect of Pe1M4, which carried a mutation unrelated to Sox binding site, was similar, but Pe1M1, in which the Sox site of Pe1 was disrupted, dropped the long promoter activity 13-fold, abolishing CEC specific enhancement from upstream elements (Figure 4A). The double mutant Pe1M1/IneM2 disrupting all the three Sox- binding sites in the short promoter decreased the long promoter activity (p<0.05) even closer to that of FO15Luc. Thus, the Sox sites in Pe1 and Ine are needed to mediate promoter activation from upstream elements. Deletion of the entire Dpe1 element or its subfragments B and C decreased the long promoter activity by 4-fold and 2-fold, respectively, in CEC culture, while deletion of Dpe2 had milder effect (data not shown).
Considering that the SI element was protected in genomic footprinting in CEC culture and bound Nfi proteins in vitro, we also mutated its Nfi contact points. Mutation SI2dm either alone or in combination with Pe1M1 or Pe1M4 dropped the long promoter activity by 10fold in CEC and similarly in other cultures indicating tissue-unspecific inhibition. Double mutation Pe1M1/SI2dm further diminished the activity (p<0.001) to the basal promoter level in mesenchymal cells suggesting an additive or synergistic effect. Thus, disruption of the Nfi site of the SI silencer element abolished both the tissue- and stage-specific promoter activity.
Dose-dependent synergy of L-Sox5/Sox6 with SOX9
Next we assessed the activation of the Matn1long promoter by cotransfected Sox proteins (Figure 2A-2D). While SOX9 doubled it, L-Sox5/Sox6 decreased the promoter activity to about half in mesenchymal cells (Figure 2A). Coexpression of L-Sox5/Sox6 with SOX9 greatly or moderately increased the ability of SOX9 to activate the promoter in CEF and HDM cultures, respectively, but rather decreased it in CEC culture. This indicates synergy between Sox proteins at early differentiation stages. When we introduced constant amount of pSOX9 and increasing amounts of pSox5 and pSox6 expession plasmids into CEF, LDM and HDM cultures, the synergistic activation peaked at low ratio of pSox5 and pSox6 versus pSOX9, and declined at elevated ratio to the level achieved by SOX9 alone (Figure 2B). Highest activation was seen in CEF (3.5-fold), followed by that in LDM and HDM cultures in inverse correlation with the endogenous Sox5 and Sox6 expression levels of these cultures (Figure 2C), raising the possibility that L-Sox5/Sox6 may modulate the activation by SOX9 in a dose-dependent manner.
We confirmed this hypothesis by forced expression of FLAG-tagged Sox trio in nonchondrocytic COS-7 cells and monitoring the protein expression in Western blots (Figure 2C). Despite the low effect of SOX9 alone, L-Sox5/Sox6 synergized with SOX9 to activate the long promoter up to ~18-20-fold at low molar excess. The activation was high from 0.9:1 to 3.6:1 molar ratio of L-Sox5/Sox6 to SOX9 in repeated experiments, but the synergy dropped above 5.3:1 molar ratio (Figure 2C). L-Sox5 and Sox6 had similar effect with sharper decline for Sox6 (Figure 2D).
By dissecting and characterizing the Matn1 DNA elements and the interacting Sox, Nfi and Hmgb1 proteins, this dissertation gives new insight into the unique control mechanism that directs Matn1expression into specific chondroblast developmental stages and distinct growth plate zones. In agreement with former reports from our group, the results presented in this dissertation uncover 1) fine tuning of the Sox9-mediated synergistic activation of the Matn1 promoter by the dose of L-Sox5/Sox6; 2) fine tuning of the Sox9-mediated Matn1 promoter activation by the dose of Nfi proteins, which peaks in early stage of chondrogenesis; 3) the dose dependent modulation of the Sox trio-mediated synergistic Matn1 promoter activation by Hmgb1.
Furthermore, based on the present and former data of our group, we proposed a model for the unique transcriptional regulation of Matn1. According to our model, the unique arrangement of the conserved DNA elements around the TATA box is an important part of this control mechanism, in which the Pe1-bound Sox9 plays a key role and mediates promoter enhancement from Dpe1. Chondroblast stage-specific regulation is achieved, because the Pe1bound Sox9 activity is finely tuned by the doses of L-Sox5/Sox6 and Nfi proteins bound to Ine and SI, respectively. The promoter activity is highest in late proliferative chondroblasts, where - due to the high expression level of chondrogenic Sox genes - the occupancy of the elements is optimal and the Ine-bound L-Sox5/Sox6 synergistically increases the transactivation by the Pe1bound Sox9, and the Sox9-mediated enhancement by Dpe1 is also optimal. In early stage of chondrogenesis, however, when the declining Hmgb1 level is higher than the raising Sox9 (Sox trio) level, Hmgb1 bound to the conserved Matn1DNA elements may fluidize the chromatin, thereby facilitating the binding of Sox9 (Sox trio) to the promoter elements and helping the activation of the gene. Nfi proteins expressed in early proliferative chondroblasts may also promote the disruption of the nucleosome structure, participate in the assembly of the PIC and contribute to promoter activation. In hypertrophic chondrocytes or under disease conditions (e.g. inflammation, tumorigenesis), the highly elevated Hmgb1 level may inhibit the promoter activity by competing with Sox proteins for their binding to the conserved Matn1 control elements.
Our model can give an explanation why the Matn1short promoter plays a critical role in restricting cartilage-specific expression and how its activity is enhanced by distal elements in transgenic mice as it was observed earlier. Remarkably, the Matn1short promoter could even restrict the activity of a powerful Sox-driven pan-chondrocytic Col2a1heterologous enhancer to distal structures and specific growth plate zones. In line with our model, the transgene carrying the IneM1 mutation displayed very lowactivity in founder embryos, but this activity remained restricted to the columnar and prehypertophic growth plate zones, as with the TR70 transgene driven by the wild-type Matn1 long promoter.
Multiple copies of the conserved Dpe1 element fused to the short Matn1 promoter directed high, zonal and distal structure-specific transgene expression resembling to that directed by the long Matn1 promoter. We concluded that Dpe1 working as an important enhancer element may account in large part for the high Sox-mediated enhancement of the Matn1 promoter in late proliferative chondroblasts for the following reasons. Dpe1 features three Sox sites binding SOX9 and L-Sox5 with opposite efficiency in vitro. It is needed for the high chondroblast stagespecific promoter activity and transactivation by the Sox trio. Dpe1 elements can exert a Sox triomediated, dose-dependent synergistic enhancement to the Matn1and Col2a1 promoters in cultures.
Our model is strengthened by the remarkable sequence and positional conservation of proximal (short) and distal DNA element observed by our collaborating partner in Debrecen (Figure 5) strongly suggesting an evolutionarily conserved transcriptional control in amniotes. Conservation of promoter and extragenic sequences in amniotes for other genes (e.g. Sox2) can reliably reflect their functional importance in development.23 Pe1 is most highly conserved among the Matn1 control elements. Sox sites of Pe1 recognized preferably by SOX9 95 bp to 195 bp upstream of TATA are most crucial for the promoter activity, but those of Ine preferably binding L-Sox5/Sox6 around the transcription start sites are also important. Sox proteins bind the two paired Sox site of Ine in a cooperative manner. Dpe1 located 18002650 bp upstream of the promoter in various species is also highly conserved in amniotes.
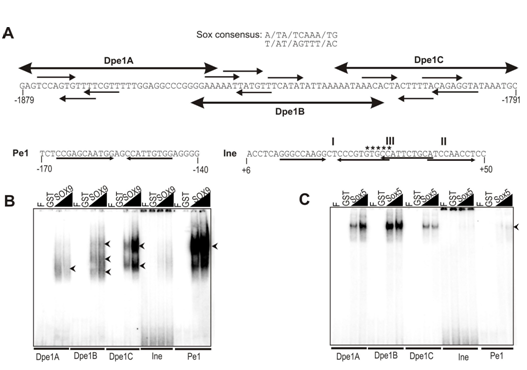
Figure 5 Comparison of binding purified GST-fused Sox proteins to the conserved DNA elements.
Figure 5A Nucleotide sequences of Dpe1, Pe1 and Ine elements. Large arrows mark the subfragments of Dpe1 and small arrows depict the Sox motifs of the elements. The conserved GTGCC motif the 5’ (I) and 3’ (II) paired Sox sites and unrelated factor-binding site (III) of Ine are denoted. Comparison of binding of GSTfused SOX9.
Figure 5B L-Sox5
Figure 5C To the Dpe1 subfragments, Ine and Pe1 in EMSA. F, free probe
Such a high degree of sequence and positional conservation among chicken and mammalian orthologs has not been found for other cartilage genes. Conserved cartilage-specific element has been identified only in the far upstream enhancer of the mammalian orthologs of Acan, but it is not conserved in amniotes. Although cartilage-specific control elements with functional Sox sites were found in varying location, e.g. in the first intron, far upstream promoter, 5’ untranslated or proximal promoter regions,21 but they do not show similarity to the Matn1control region.
The transcriptional regulation of the Matn1 involves similar as well as different molecular mechanism as compared to other cartilage protein genes. The Sox trio likewise plays essential roles in transcriptional regulation of other cartilage-specific genes,21 but Matn1 is regulated differently by Sox trio than other cartilage ECM genes. One difference is that whereas Sox9 is sufficient for the activation of Col2a1, AcanandCrtl1, L-Sox5/Sox6is also absolutely required for the activation of Matn1, as Matn1 mRNA was not detected in Sox5-/-;Sox6-/-mice.18 Furthermore, we found that L-Sox5/Sox6 modulates the Matn1 promoter activation by SOX9 in a dose-dependent manner. This effect is likely due to unique set of conserved DNA elements that are capable of interacting with Sox proteins with different efficiency.
In addition to the role of Sox trio, we also demonstrate the novel role of Nfi proteins in the gene regulation. We provide evidence that Nfi proteins also modulate the promoter activity and the SOX9-mediated trans activation in a dose-dependent manner. They increase the activation by SOX9 at low dose, but repress that at high dose. In agreement with the transient activation of Nfi genes during in vitro chondrogenesis in QRT-PCR, dominant negative mutation of Nfib interfered with chondrogenesis, while over expression of the wild type Nfib increased the Sox9 and Col2a1 expression,24 Nfi sites mediating this regulation however have not been identified yet. By extending novel results and previous data from our laboratory (EMSA, in vivo footprinting), we provide evidence that, in addition to the Sox trio, Nfi proteins binding SI near TATA also play critical role in fine tuning and enhancement of the chondroblast stagespecific activityof theMatn1promoter. Based on genomic footprinting, the Nfi motifs of SI and SII are first occupied by in vivo bound transcription factors (Figure 4), in line with the transient Nfi expression in early chondrogenesis.
Our model is consistent with former observations that in vivo footprints were absent from the Matn1 short promoter in the non-expressing CEF and they gradually appeared in differentiating HDM culture, strongly suggesting that activation of Matn1 involves regulation at chromatin level. In fact, the Nfi sites of SI and SII were not occupied in vivo in CEF, albeit Nfi genes are expressed in CEF (Figure 4D) and Nfi proteins can bind SI and SII from CEF extracts in EMSA and in genomic footprinting. Based on their interaction with histones and GTFs,25,26 we can assume that Nfi proteins may help to disrupt the nucleosome structure and contribute to Matn1 activation. The unique molecular mechanism described in this dissertation may facilitate the construction of growth plate zone-specific vectors and the development of biotechnological therapies for skeletal diseases.
None.
None.

©2015 Moshiri. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.