MOJ
eISSN: 2373-4442


The emergence of the pandemic HINI influenza virus in humans in early April 2009 in Mexico and California came as a total surprise. A total of 31,837 cases positive for Influenza A H1N1 and 1551 confirmed deaths were reported till 11th May 2010 by Government of India. Viral infections are in general self-contained after activation of the innate immune defense. TNF-α is a potent proinflammatory cytokine produced early in the innate immune response to viral infection that promotes a wide range of immunologic responses. Excessive systemic TNF-α is responsible for many symptoms of clinical infection and may lead to fatal complications. Individuals differ with respect to the level of cytokine production after the virus infection and that these differences may be attributed partly to single nucleotide polymorphism (SNPs) within its corresponding gene. Thus, the present study was planned to investigate the role SNPs in the TNF-α promoter gene in 2009 H1N1 positive patients in India.
Genomic DNA was isolated from 30 Influenza A 2009 pandemic H1N1 infected patients and 20 healthy controls. SNPs of -238G/A, -308G/A, -857C/T and -863C/A were assessed by polymerase chain reaction of TNF-α promoter gene followed by restriction fragment length polymorphism.
Although the number of 2009 pandemic H1N1 infected patients carrying -308A and -238A alleles were observed to be more than that of the controls but no statistical difference was observed in the genotypic and allelic frequency distribution of all TNF-α promoter loci between the cases and controls. However, further studies involving larger number of Indian patients, are required to substantiate the present finding.
Keywords: influenza, H1N1, pandemic, TNF-α, polymorphism, India
ARDS, adult respiratory distress syndrome; ALI, acute lung injury; SNPs, single nucleotide polymorphism; rRT-PCR, recommended real-Time RT-PCR; NTC, no template control
The emergence of a new influenza A (H1N1) virus in early April 2009 in Mexico and California, quickly spreading worldwide through human-to-human transmission, generated the first influenza pandemic of the 21st century. India reported its first case on 13th may, 2009. According to the Directorate General of Health Services, Government of India, a total of 31,837 cases positive for Influenza A H1N1 and 1551 confirmed deaths were reported till 11th May 2010. The virus was reported to be antigenically unrelated to the seasonal influenza viruses but molecular analysis of the new influenza A (H1N1) 2009 pandemic virus genome showed that it was derived from several viruses which have been circulating in pigs for years, namely the North American H3N2 triple- reassortant, the classical swine H1N1 lineage, and the Eurasian ’avian-like’ swine H1N1 virus.1
The infection was rather self limiting, with symptoms of illness of the upper respiratory tract. The ubiquitous characteristic of this H1N1 pandemic compared with other influenza A pandemics was the occurrence of more severe cases among young individuals. Severity was defined by the advent of pulmonary complications, namely pneumonia, acute lung injury (ALI), and adult respiratory distress syndrome (ARDS).These were also involved in pregnant women and among individuals with underlying diseases such as obesity, bronchial asthma, and heart failure.2
The host immune responses have been implicated in contributing to severe respiratory pathogenesis of H1N1 infections. Tumor necrosis factor- α is a pro-inflammatory cytokine, which has been referred to a “mixed blessing for the higher organism”.3,4 Although the controlled self-limited expression of TNF plays a critical role in activating host defense mechanisms but uncontrolled over expression of TNF accounts for the devastating consequences for the host organism, consequently leading to diffuse inflammation, multiorgan dysfunction, hemodynamic collapse, cardiomyopathy and variety of other diseases.5 Recent studies have found that levels of circulating TNF-α are elevated in various infectious disease such as Hepatitis, Japanese encephalitis, avian influenza etc.6,7 Although the exact clinical significance of these findings is uncertain, it has been postulated that that individuals differ with respect to the level of cytokine production after the virus infection and that these differences may be attributed partly to single nucleotide polymorphism (SNPs) within its corresponding gene.
Under circumstances where the release of TNF-α has been triggered the genetically endowed capacity for greater TNF-α production leads to severe inflammatory reactions. Till date 621 SNPs in TNF- α gene have been reported in the NCBI website , with more than 10 SNPs in the promoter region, including -238 G/A , -244 A/G, -308 G/A, -376 A/G, -575A/G, -857C/T, -863 C/A, -1031T/C.8,9 Regarding the TNF gene, four SNPs have been well associated with the higher TNF-α production after viral infections,6,10 all at different locations within the promoter region: a substitution of guanine by adenine at the -238 position(-238 TNF G/A), a substitution of guanine by adenine at the -308 position(-308 TNF G/A), a substitution of cytosine by thymine at the -857 position(-857 TNF C/T), a substitution of cytosine by adenine at the -863 position(-863 TNF C/A).
It has been observed that the swine flu H1N1 virus causes more severe pathological lesions in the lungs of infected mice, ferrets and non-human primates than the seasonal influenza virus.11 Also, a study conducted by Bermejo et al. has shown that Th1 responses (IFNγ, TNF-α, IL-15) were elicited in severe patients infected by H1N1 patients.12 Therefore, to determine if host immune responses play a potential role in the pathogenesis of H1N1 infected patients the present study investigated the role of SNPs in the TNF-α promoter gene in 2009 H1N1 positive patients in India.
Study subjects
Thirty 2009 Influenza A pandemic H1N1 (Inf A pdm H1N1) infected patients confirmed by reverse transcriptase RT-PCR (rRTPCR) were included as the study group whereas twenty age and sex matched asymptomatic healthy individuals were included as controls.
Throat swab and nasal swab were collected from all the cases (n=30) and controls (n=30). Swabs were transported in Viral Transport Medium (VTM HiMedia, Mumbai, India) to the laboratory in cold chain. A part of the sample was subjected for H1N1 testing by Real Time RT-PCR (rRT-PCR) and the remaining was stored at -70°C till further use.
The samples were collected from the suspected H1N1 cases for laboratory confirmation by real time PCR as Department of Virology was one of the referral H1N1 testing centers appointed by Govt. of India. The study protocol was approved by Institute's Ethics Committee.
Confirmation of Influenza A pandemic H1N1
All the samples collected from the suspected 2009 Inf A pdm H1N1 infected patents were handled in the Class II Biosafety cabinet with Class III Biosafety containment protective measurements. Viral RNA was extracted from 140µl of sample using RNeasy Minikit (QIAGEN, GmbH, Hilden, Germany) according to the manufacturer’s instructions.
The CDC recommended Real-time RT-PCR (rRT-PCR) protocol for detection and characterization of 2009 Inf A pdm H1N1 was used for the in-vitro qualitative detection and characterization of swine influenza viruses in respiratory specimen.13 The influenza A primer and probe set is designed to specifically detect North American swine influenza A viruses. The pandemic H1 primer and probe set is designed to detect Influenza 2009 A/H1N1 pdm virus H1 influenza. Human RNase P gene is used as housekeeping gene whereas nuclease free water in place of RNA input was used as No Template Control (NTC).
The samples positive for Influenza A, SwA and SwH and housekeeping RNAase P genes were confirmed as Inf A pdm H1N1 positive.
TNF-α promoter polymorphism by PCR-RFLP
Respiratory samples were centrifuged at 5000rpm for 10 minutes to pellet down the cellular materials. Supernatant was removed and genomic DNA was extracted from the pelleted cellular materials using the DNA extraction kit (QIAGEN, GmbH, Hilden, Germany) as per the manufacturer's protocol and stored at -20°C until further use.
The SNPs of -238G/A, -308G/A, -857 C/T and -863C/A in TNF-α gene were detected by the gene-specific PCR followed by restriction fragment length polymorphism (PCR-RFLP). The PCR was carried out using the specific primers as described earlier.14 PCR products were further subjected to restriction digestion by allele specific restriction enzymes by overnight incubation (16-18 hours) at 37°C.14 The digested products were separated by 12% Polyacrylamide gel electrophoresis followed by ethidium bromide staining and evaluated after comparison with appropriate controls.
Statistical analysis
The data was analyzed using SPSS software version 16.0. The TNF- α promoter gene polymorphism in cases and controls was evaluated using x2 SPSS was used to calculate odds ratio (OR) and 95% confidence intervals (CI) for assessing the risk associated with particular allele and genotype. The ‘P value’ of <0.05 was taken as significant.
The present study included 30 confirmed Influenza A Pandemic H1N1 cases and 20 apparently healthy individuals as controls. The age of both cases and controls was in the range of 15-65 years. The male to female ratio in test group was 1:1 and 1:3 in controls respectively.
DNA of all the cases and controls was subjected to amplification of TNF-α gene promoter region -238G/A, -308 G/A, -857 C/T and -863 C/T. The amplified products obtained for the four positions were 152bp, 143bp, 131bp, and 133bp respectively (Figure 1).
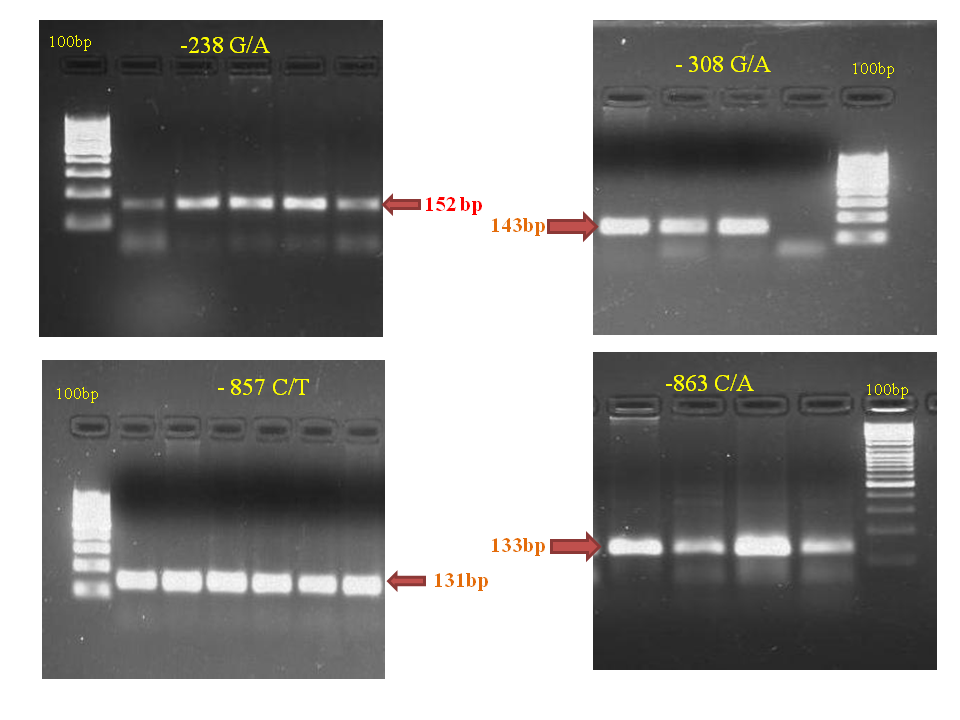
Figure 1 Agarose gel electrophoresis of PCR products of the TNF-α gene -238, -308, -857 and -863 regions.
These PCR products were further subjected to restriction digestion. The genotypic and allelic frequency of TNF-α polymorphism at all the four positions -238G/A, -308G/A, -857C/T and -863C/A between the cases and controls is described in Table 1.
Loci |
Genotype |
Cases (N=30) |
Controls (N=20) |
238 |
GG |
(27/30)90% |
(20/20)100% |
GA |
(2/30)6.6% |
(0/20)0 |
|
AA |
(1/30)3.3% |
(0/20)0 |
|
308 |
GG |
(16/30)53.3% |
(12/20)60% |
GA |
(8/30)26.6% |
(8/20)40% |
|
AA |
(6/30)20% |
(0/20)0 |
|
863 |
CC |
(19/30)63.3% |
(9/20)45% |
CA |
(8/30)26.6% |
(9/20)45% |
|
AA |
(2/30)6.6% |
(2/20)10% |
|
857 |
CC |
(24/30)80% |
(14/20)70% |
CT |
(4/30)13.3% |
(5/20)25% |
|
TT |
(2/30)6.6% |
(1/20)5% |
Table 1 Genotypic and allelic frequencies of TNF-α polymorphism
The different statistical parameters of genotypic and allelic frequency of TNF alpha promoter polymorphism between cases and control groups is shown in Table 2. No statistical difference was observed in the genotypic and allelic frequency distribution of all TNF-α promoter loci between the cases and controls.
Loci |
Genotype |
H1N1 Positive Cases vs Controls |
|||
X2Value |
P value |
Odds Ratio |
95% CI |
||
238 |
GG |
2.128 |
0.265 |
0.574 |
0.449-0.735 |
GA |
1.389 |
0.51 |
1.714 |
1.350-2.177 |
|
AA |
0.68 |
1 |
1.69 |
1.339-2.132 |
|
308 |
GG |
0.216 |
0.773 |
0.762 |
0.242-2.398 |
GA |
0.535 |
0.548 |
0.643 |
0.196-2.108 |
|
AA |
4.545 |
0.069 |
1.833 |
1.400-2.401 |
|
863 |
CC |
1.637 |
2.51 |
2.111 |
0.667-6.682 |
CA |
1.797 |
0.229 |
0.444 |
0.134-1.470 |
|
AA |
0.181 |
1 |
0.643 |
0.083-4.981 |
|
857 |
CC |
0.658 |
0.506 |
1.714 |
0.463-6.351 |
CT |
1.107 |
0.454 |
0.462 |
0.107-1.988 |
|
TT |
0.059 |
1 |
1.357 |
0.115-16.047 |
|
Table 2 Statistical comparison of Genotypic and allelic frequencies of TNF-α polymorphism between cases and control groups
Acute respiratory distress syndrome is one of the major causes of mortality in pandemic Influenza A H1N1 infection. Analysis of the immune mediators involved in host responses to the virus may elucidate the pathogenic events leading to poor outcomes. Th1 (IFN-γ, TNF-α, IL-15) and Th-17 (IL-8, IL-9, IL-17, IL-6) cytokine responses are reported as hallmark of respiratory compromise following infection of H1N1.12 These cytokines not only promote antiviral immunity but also contribute to inflammation of respiratory tract by recruitment of neutrophils and mononuclear cells to the site of the infection.
Circulating TNF-α levels and their genetic associations between gene polymorphisms have been established in many diseases. Recent studies have elucidated the genetic alterations in the TNF-α locus are involved directly in high TNF-α production. Several polymorphisms have been identified inside the TNF-α promoter positioned at (relative to the transcription start site) -1031(T/C), -863(C/A), -857(C/A), -851(C/T), -419(G/C), -376(G/A), -308(G/A), -238(G/A), -162(G/A), -49(G/A). However, significance of the -238(G/A), -308(G/A), 857(C/A), -863(C/A) alleles of TNF-α gene with the susceptibility to H1N1 infection is neglected entity so far.
Several studies have investigated the association between TNF alleles and viral infections. The 238*A allele was associated with chronic infection with hepatitis B virus.9 Study conducted by Pujhari et al. showed that disease severity and disease progression of Japanese encephalitis was associated with the 308 and 863 alleles of patients.6
The single nucleotide mutation at -238 and -308 positions of TNF-α promoter has been found to predispose to higher TNF production.15 A point mutation from C to T at -857 possessed higher transcription due to the change in the binding stability by variant allele whereas polymorphism at position -863C/A in the promoter region has been reported to be associated with reduced TNF-α promoter activity and lower plasma TNF levels.10,16
Regarding the TNF gene, four SNPs -238, -308, -857 and -863 have been well associated with viral infections. Thus, carriage of these SNPs by humans may affect their susceptibility to H1N1 infection and can be responsible for severe pulmonary complications in young individuals. Considering this fact in present study these four SNPs in 2009 Influenza A pandemic H1N1 positive Indian patients were compared with the healthy individuals of the same region. Although, number of H1N1 patients carrying -308A and -238A alleles were observed to be more than that of the controls (Table 1) but no statistical significance was observed. This could be due to the small number of enrolled patients compared with the high frequency of 2009 H1N1 influenza cases as well as the geographical variation.
So far in only one study conducted by Antonopoulou et al.,17 in Greece, role of -238, -308, -376 SNPs in 109 H1N1 infected patients was studied. -238A allele was observed to be significantly higher in 2009 H1N1 cases than controls and carriage of -238A allele was associated with the development of pneumonia.17 However, these results are specific to a geographic region and have to be confirmed in different cohort.
Since, this was a pilot study statistical significance could not be established thus, further studies with larger number of Indian H1N1 infected patients are needed to be done to prove the present results statistically.
The present pilot study showed no significant difference in TNF-α promoter gene polymorphism between Pandemic Influenza A H1N1 infected Indian patients and healthy individuals. However, further studies involving larger number of Indian patients, are required to substantiate the present finding.
None.
There are no financial conflicts of interest.
None.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.