MOJ
eISSN: 2575-9094


Research Article Volume 2 Issue 4
1Department of Microbiology, Dolphin (PG) College of Science and Agriculture, India
2Department of Microbiology, Punjab University, India
Correspondence: Vikas Pahal, Department of Microbiology, Dolphin (PG) College of Science and Agriculture, India, Tel +91-9872193964
Received: July 11, 2018 | Published: July 27, 2018
Citation: Pahal V, Devi U, Dadhich KS. Quercetin, a secondary metabolite present in methanolic extract of Calendula officinalis, is a potent inhibitor of peptide deformylase, undecaprenyl pyrophosphate synthase and DNA primase enzymes of Staphylococcus aureus: an in vitro and in silico result analysis. MOJ Drug Des Develop Ther. 2018;2(4):216-225. DOI: 10.15406/mojddt.2018.02.00050
Plant‒based naturally occurring phytochemicals have great inhibitory potential against various pathogenic bacteria, but the mechanism of inhibition and the essential cell molecules to which they bind and inhibit, remain unclear and unexplored so far. In the present study, we examined the effect of secondary metabolites from C officinalis both in vitro and in silico to decrypt the probable pathway of inhibition. In in vitro experiments, Staphylococcus aureus was used to evaluate the antibacterial potential of secondary metabolites present in different organic extracts of C officinalis using agar‒well diffusion and XTT‒colorimetric methods. In in silico experiments, 4 important C officinalis secondary metabolites viz. Alpha cadinol, Scopoletin, Esculetin and Quercetin were docked against 4 essential enzymes of bacteria like Peptide deformylase, Gamma hemolysins, Undecaprenyl pyrophosphate synthase and DNA primases to assess their antibacterial potential using AutoDock Vina.1 The enzyme‒ligand interaction of docked complexes was further analyzed by Molecular Dynamics Simulation technique using GROMACS (4.6.6). As per in vitro results, methanolic extract was found to be the most promising extract having highest antibacterial potential, where the 20mg/ml concentration of the extract was found to completely inhibit the bacterial growth, whereas for pure Quercetin the complete inhibition was achieved at 5mg/ml concentration. MIC was observed to be 20 and 5mg/ml with methanolic and pure Quercetin extracts, respectively. Whereas, MBC values were found to be 40 and 5mg/ml for methanolic extract and Quercetin, respectively. As per in silico results, Quercetin was found to be the only secondary metabolite which strongly inhibited three essential enzymes of S aureus like Peptide deformylase, Undecaprenyl pyrophosphate synthase and DNA primases enzymes, which resulted in the inhibition of post‒translation, cell‒wall biosynthesis and initiation of replication pathways, respectively. As Quercetin was found in methanolic extract of C officinalis, we have successfully demonstrated that C officinalis has great potential to inhibit the growth of S aureus.
Keywords: antibacterial, phytomedicines, docking, molecular simulation
CFU, colony forming unit; DMSO, dimethyl sulphoxide; OD, optical density; PDB, protein data base; PS, picoseconds; SD, standard deviation; ZOI, zone of inhibition
Pathogenic bacteria are responsible for deterioration of human health which is a major concern worldwide and especially in the developing countries like India. Secondly, irrelevant and irrational use of antibiotics and other synthetic antimicrobial agents has put strong selective pressure on various pathogenic bacteria turning them into resistant strains, which has created an unparalleled challenge before scientific communities to combat against these resistant pathogenic microbes.1 This triggers the need to develop novel and active alternative remedies like plant‒based medicines, which are already used in folk medicines throughout the world, but have not been authenticated pharmacologically. Plant based drugs are economical and have great antibacterial potential as they simultaneously mitigate many side effects of synthetic antimicrobials agents.2 Plants produce secondary metabolites in a very low concentration in normal conditions of developmental and physiological stages, but the production increases many folds under various abiotic and biotic stresses like microbial and other pathogenic attacks. These secondary metabolites like flavonoids, tannins, alkaloids, phenols/polyphenols, volatile essential oils, terpenes, coumarins etc. are the defense molecules of plants and they possess unique and complex chemical structures which are very diverse in nature. These complex and unique biochemical structures have well documented antibacterial properties and variously used as agrochemicals, biopesticides and flavor and fragrance ingredients.2,3 So, plant species are a potential reservoir for the discovery of new antimicrobial drugs. For the last many decades, these plant‒derived chemicals have successfully been used against many microbial infections which have created an upsurge of interest in the antimicrobial potential of medicinal plants. It was estimated that nearly 40% of compounds used in pharmaceutical industry are directly or indirectly derivatives of plants because the chemical synthesis of such compounds is either not viable or economically impracticable.4‒8 But, the mechanism of inhibition and the essential cell molecules to which these phytochemicals bind and inhibit various bacterial essential biochemical pathways still remain unexplored.9 Study of these mechanisms of bacterial inhibition by phytochemicals is absolutely imperative because it helps the scientists in developing novel improved antimicrobial molecules using various in silico techniques.
Staphylococcus aureus is a gram‒positive bacteria which is a leading cause of hospital‒associated and community‒associated infections worldwide. S aureus have the inimitable ability to escape from both the innate and the adaptive immune response by using multiple intrinsic virulence factors and pathways which can incapacitate various immune system molecules which results into improper or failed immune response.10 S aureus infection is of greatest concern because of its intrinsic virulence, ability to cause varied range of life‒threatening infections, and ability to adapt different environmental conditions which resulted in evolution of resistant strains like MRSA (methicillin‒resistant S aureus) strains.11 Calendula officinalis Linn (commonly known as “African marigold”) belongs to the family Asteraceae and is used in folk medicines mainly in Europe, China and India and also in other regions of the world. This plant has well documented multiple pharmacological activities such as anti‒HIV, anti‒inflammatory, hepatoprotective, spasmolytic, spasmogenic, antibacterial and antifungal.12 Chemical and pharmacological studies show the presence of various classes of phytochemicals like flavonoids, triterpenoids, quinones, coumarines, volatile and essential oils, carotenoids and others. These secondary metabolites are taken to be responsible for its medicinal properties.13 In the present paper, we tried to decode the probable pathway by which some secondary metabolites from C officinalis inhibit the pathogenesis of S aureus infection by obstructing the action of various important enzymes. The project was accomplished in two parts: In the first part, known as in vitro experiments, various active constituents of C officinalis were isolated using different organic solvents like hexane/ethyl‒acetate and methanol. The bactericidal effects of these various organic extracts and of pure Quercetin (Phytochemical found in various medicinal plants including C officinalis were analyzed using two methods: agar‒well diffusion method and XTT‒colorimetric method. In Second part, known as in silico experiments, various bioinformatics softwares and programmes were used to find out the probable mechanism of inhibition of S aureus by various phytochemicals present in different organic extracts of C officinalis.
In vitro experiments
In vitro experiments were performed to reconfirm the antibacterial effect of hexane/ethyl‒acetate and methanolic extracts of C officinalis and the extracts were further analyzed biochemically for the presence of various medicinally important phytochemicals.
Chemicals
All the chemicals used in the present study were of analytical grade. Nutrient agar and broth were purchased from Hi Media Pvt. Ltd, India. Menadione, XTT‒salt and Quercetin were purchased from Sigma‒Aldrich, India. Organic solvents like hexane, ethyl‒acetate and methanol were purchased from Merck, India.
Collection and processing of leaves and flowers
The leaves and flowers of C officinalis were collected and washed with sterile double distilled water. Washed material is then air dried at room temperature (35‒40°C) for 6‒7days and homogenized to a fine powder using a sterilized mechanical grinder. This coarsely powdered material was first extracted with hexane and ethyl acetate (50:50v/v) for 72hrs to isolate the essential oils and other fatty acids using Soxhlet assembly. After this, the defatted material was used for cold‒percolation method to isolate the active principles soluble in methanol. Methanol is considered as the most suitable solvent for the isolation of phenolic and flavonoid compounds. Methanolic Preparation was filtered through a sterilized filter paper (Whatman No. 1) and the filtrated solvent was evaporated to dryness under vacuum at 40‒50°C using a rotary evaporator. The dried extracts were then sterilized by UV‒irradiation (30‒40minutes with regular shuffling), checked for sterility on nutrient agar plates and stored at 4°C in sterile glass bottles until further use.9
Test microorganism
Gram‒positive bacterial strain of Staphylococcus aureus (MTCC 737) was obtained from Institute of Microbial Technology (IMTECH), Sector 39, Chandigarh, U.T, India.
Screening for antimicrobial activity
S aureus was grown overnight in nutrient broth at 37°C for 18‒20hrs. Midlogarithmic phase organisms were reaped by inoculating this culture further into 30ml of fresh broth for additional 2.5 to 3.5hrs at 37°C. The bacterial pellets were collected from the broth culture after centrifugation at 1000g for 10minutes at 4°C. These bacterial pellets were washed and resuspended in cold 10mM sodium phosphate buffer (pH=7.4). The optical density of bacterial aliquot was measured at 620nm and the concentration of bacteria was standardized (OD620 0.20=5X107CFU/ml).14 Antimicrobial studies were carried out using cup (well) assay method also known as agar well diffusion method. In this method, approximately, 20‒25ml of pre autoclaved agar media cooled at 45°C was poured into petri plates and allowed to solidify at room temperature. One hundred microliter (100μL) of the inoculum of S aureus was spread onto the agar plates so as to attain a confluent growth. Then four wells were made with a sterile borer in the inoculated agar plates. Initially, stock concentration of 40mg/ml was prepared with 8% DMSO and different concentrations of extracts were made by serial dilution. The concentrations used in agar well diffusion method were 10mg/ml, 5mg/ml, 2.5mg/ml and 1.25mg/ml. In case of Quercetin, concentrations which were used for study of antibacterial activity were 5.0mg/ml, 2.5mg/ml, 1.25mg/ml and 0.62mg/ml. A 100μL volume of each extract was propelled directly into the wells of the inoculated agar plates. The plates were allowed to stand for 1‒1.5hour at room temperature for diffusion of the extract into agar and further incubated at 37°C for 18‒24hours. 8% DMSO served as the negative control and antibiotic erythromycin served as the positive control. The experiments were performed in triplicates and the mean values of the diameter of Zone of Inhibition (ZOI) were calculated in millimeter scale with standard deviations.9
XTT‒colorimetric method for measuring antibacterial potential of extracts
Antibacterial activity with different concentrations of plant extracts and Quercetin was also evaluated by the XTT‒colorimetric method.15 This is an indirect method which measures the antibacterial activity by calculating the Electron Transport System (ETS) activity by means of redox dyes (artificial electron acceptors) that can successfully compete with oxygen for electrons. XTT is a tetrazolium salt (2, 3‒bis [2‒methyloxy‒4‒nitro‒5‒sulfophenyl] ‒2H‒tetrazolium‒5‒carboxanilide) which is reduced by the dehydrogenase enzymes of ETS system to a water soluble formazan dye, and absorbance of which can be measured colorimetrically at 490nm. The measurement of relative reduction in formazan overall provides an approximation of antimicrobial activity of respective phytochemical. Concisely, overnight broth culture of S aureus was adjusted with nutrient broth to attain the standard concentration of bacteria (5X107CFU/ml) and 170µl of this adjusted broth cultures were added to 96‒well flat‒bottom plate. 30µl of different concentration of plants extract (concentration ranges from 20mg/ml to 0.64mg/ml) were added to the well with gentle mixing, and incubated for 15hours. In case of Quercetin, concentrations which were used for study of antibacterial activity were from 10mg/ml to 0.64mg/ml. On succeeding day, 100µl of each well material was transferred into new flat‒bottom plate. Fresh solution of XTT+menadiaone was prepared and 25µl of that solution was added to each well with gentle mixing. Plates were incubated for 1 hour at 37°C and the reading was taken at 490nm with the help of Plate‒reader. Negative control was the media containing S aureus without any growth inhibitor and positive control was the media with bacteria and antibiotic erythromycin (2mg/ml). Final readings were adjusted after deducting the reading of various blanks.9 Experiment was performed in triplicates and final reading was taken as mean (±SD). Antimicrobial activity was measured as percentage reduction (% Rd) of bacterial growth with the following formula:
Assay for MIC and MBC
Minimum Inhibitory Concentration (MIC) for S aureus was determined by microdilution broth method (XTT‒calorimetric) as described earlier. The lowest concentration that did not permit any growth, as confirmed by the OD of the plate was considered as MIC. Maximum Bactericidal Concentration (MBC) is the lowest concentration of antimicrobial agent that will not allow the growth of an organism after sub‒culturing on antibiotic free nutrient agar media.16 MBC was determined by sub‒culturing the preparations that did not show any bacterial growth (as per the results of MIC). A 100μL aliquot from the selected tubes (showing complete inhibition as per MIC results) was spread over the nutrient agar plate and incubated at 37°C for 24hours and examined for bacterial growth, if any.
Test for phytochemicals
Standard protocols were used to find out the presence of phenols, flavonoids and coumarins in hexane/ethyl‒acetate and methanolic extracts of C officinalis.1,2,6,9
Test for Phenols
First test (Gelatin test): 1ml of 1% gelatin solution (having 10% NaCl) was added to extracts. Formation of precipitate indicates the presence of phenolic compounds.
Second test: Methanolic extracts were first dissolved in 5ml water followed by addition of few drops of neutral ferric chloride solution. Formation of dark green colour indicates the presence of phenolic compounds in the extracts.
Third test: Extracts were treated with aqueous ammonia. In presence of air, green colour formation indicates the presence of phenolic compounds.
Test for flavonoids
Extracts were mixed with 2ml of 50% methanol solution. After warming the solution a small piece of magnesium metal was added and after that concentrated HCL was added dropwise. Occurrence of crimson to magenta colour shows presence of flavanones, orange colour indicates presence of flavones and red colour indicates presence of flavonols.
Test for coumarin
10% NaOH was added to various extracts followed by addition of few drops of chloroform. Observation of yellow colour indicates the presence of Coumarin.
Four potential phytochemicals were tested against 4 important bacterial enzymes using various bioinformatics softwares and programs. These phytochemicals were reported to be present in various organic extracts of Calendula officinalis.
Selection and properties of phytochemicals
Four phytochemicals of Calendula officinalis viz. Alpha cadinol, Scopoletin, Esculetin and Quercetin were selected for drug docking and MD simulation studies. Structures of these ligand molecules Figure 1 were retrieved from PUBCHEM server (https://pubchem.ncbi.nlm.nih.gov/). “Lipinski’s rule of five” was used to screen the phytochemicals in the ISIS BASE and they were found to display significant drug‒like properties such as number of H‒bond donors ≤5, number of H‒bond acceptors ≤10, molecular weight ≤500, high lipophilicity (logP) ≤5, etc. Other details of these molecules are shown in Table 1. These selected bioactive compounds were then drawn in Molinspiration (http://www.Molinspiration.com/cgi‒bin/properties) to predict their bioactivities and molecular properties. The 3D structures of the selected compounds were drawn and the corresponding pdb files were downloaded using ChemSketch (http://www.acdlabs.come) programme and subsequently were viewed using PyMOL window (http://www.pymol.org).17‒19
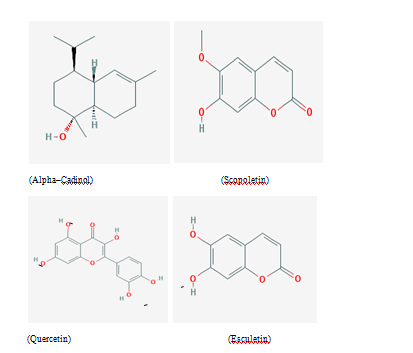
Figure 1 Chemical structure of phytochemicals used in the present study for docking and MD simulation studies.
Phytochemicals |
Hydrogen bond (donor) |
Hydrogen bond (acceptor) |
Lipophilicity (logP) |
Aqueous solubility (logS) |
Molecular weight |
Alpha cadinol |
1 |
1 |
4.32 |
‒3.09 |
222.2 |
Scopoletin |
4 |
1 |
1.43 |
‒2.53 |
192 |
Esculetin |
2 |
4 |
1.08 |
‒2.42 |
178.03 |
Quercetin |
5 |
7 |
2.11 |
‒3.87 |
302.04 |
Table 1 Physico‒chemical properties of various phytochemicals used in the present study
Alpha‒cadinol (C15H26O)
α‒cadinol (Cadin‒4‒en‒10‒ol), was found to be present in essential oils of many medicinal plants. It belonged to Sesquiterpenes family of oil and has well documented antibacterial and antifungal activity against pathogenic strains of gram‒positive bacteria (S aureus, Bacillus spp.), gram‒negative (E coli, S typhi, K pneumoniae, P mirabilis) bacteria and against fungus C albicans.20
Scopoletin (C10H8O4 ) and esculetin (C9H6O4)
Scopoletin (7‒hydroxy‒6‒methoxychromen‒2‒one) and Esculetin (6,7‒dihydroxychromen‒2‒one) belong to phenolic coumarin class of phytochemicals. Both of these phytochemicals get accumulated in plants in response to mechanical injury, microbial attacks and even due to other stress. These phytochemicals are responsible for inhibitory effects on various microbes and were found in methanolic extract of plants.13,21
Quercetin (C15H10O7)
Quercetin (2‒(3,4‒dihydroxyphenyl)‒3,5,7‒trihydroxychromen‒4‒one) is a naturally occurring polyphenolic flavonoid and was found in methanolic extract of medicinal plants. It is ubiquitous in plant kingdom and a major bioflavonoid in the human diet. This phytochemical possess profound antibacterial, antifungal activities, antioxidant, anti‒inflammatory and anticancer activities.22‒24
Retrieval of protein/enzyme sequences and homology search
The retrieval of protein sequences (with their accession number and PDB Id) for four target enzymes i.e DNA primase (ABR52502, 4E2K_A), Gamma hemolysin (BAA07714, 3B07_B), Undecaprenyl pyrophosphate synthase (ABR52197, 4H8E_A) and Peptide deformylase (KII21618, 1WS0_A) of S aureus were retrieved from National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/). BLASTp was performed to find similarity search for four target enzymes. After analyzing similarity, protein modelling for all the four enzymes targets was done using Modeller 9.15. Then, on the basis of lowest Discrete Optimized Protein Energy (DOPE) score, best model was selected for all the four targets. Further, the quality of the target model was evaluated using Ramachandran plot and Prosa web server. After the analysis, it was observed that all the enzymes residue were present in the allowed region of the Ramachandran plot and it constitute the enzymes as of good quality target for docking and molecular simulation studies.9,17
Molecular docking analysis
The process of docking involves the binding of drug molecule to the active site of its enzymes. The active sites present in the target enzymes were identified using Ligsite (http://projects.biotec.tu‒dresden.de/pocket/) programme, which is used for the automatic detection of various pockets (binding sites for phytochemicals) on the surface of target enzymes. Auto Dock Vina, which is based on Lamarckian genetic algorithm (LGA), was used to scan enzyme‒ligand interactions and the process was completed by assigning a binding energy to each conformation.18 In the subsequent steps, polar hydrogen was added and the protein molecules were assigned with Kollman united atom charge followed by generation of the respective PDBQT file. The 3D affinity grid fields were created using autogrid programme with grid map of 40°A×40°A×40°A points, which encloses the entire groove near the active site to fit the ligand. The finest docking results were always the one having lowest energy.25 Amino acids residues within the circumference of 4.0A° around the molecule in the molecular docking were represented as active sites for binding. The binding free energy between the active site and ligand was calculated using Calculate Interaction Energy (CIE) programme. Those amino acids whose absolute values of interaction energy (Vander Waals forces & electrostatic energy) with ligands were above 2.5kcal/mol were considered as key molecules and selected for further study.18 The structural models of targeted enzymes were constructed by Discovery Studio (www.accelrys.com). LigPlot+ (V.1.4.5) programme was used to drive LigPlots of the docked complexes and to extract unambiguous images of the hydrophobic interactions pertaining to amino acid residues in the enzyme under study.25,26
Molecular dynamics simulation and analysis of molecular dynamics trajectory
MD simulation analyzes molecular motion on the atomic scale as a function of time. To evaluate the steadiness of enzyme‒ligand complex, simulation was performed on Ubuntu (14.04) operating system using command‒line based software GROMACS (4.6.6).27 The lowest binding energy (most negative) as per docking analysis generated by AutoDock programme was considered as initial conformation for MD simulation. The molecules were placed in the center of a dodecahedron box filled with water molecules, and Na+ ions were added in each box to neutralize the system. After standard equilibration procedure a MD cycle was run and potential energy of enzyme and stability of the binding was evaluated. Enzyme‒ligand complexes were energy minimized by steepest descent of 5000steps. Whole system was exposed to position‒restrained dynamics simulation where modified berendsen thermostat coupling scheme algorithm was employed for both NPT (constant number of atoms N, pressure P and temperature T) and NVT (constant number of atoms N, volume V and temperature T) ensemble at 300K for 100ps to equilibrate the system. Finally, the whole system was subjected to the MD run at 300K temperature and 1 bar pressure for simulation time of 50,000ps. All the trajectory files were explored using GROMACS utilities, recorded and stored for further analysis. Energy fluctuations and RMSD (root‒mean‒square deviation) of the complex in each trajectory were analyzed with respect to simulation time. The numbers of hydrogen bonds formed during the simulation were calculated using ghbond utility and therefore used to reconfirm the docking results in terms of hydrogen bonding pattern between the receptor and the ligand.28 It uses a default distance cutoff of 3.5A° (0.35nm) and a default bond angle cutoff of 300 or greater. XMGRACE programme was used to analyze the trajectories results and respective graphs were plotted. Functions and association of molecules in a biomolecular structure, their stability and flexibility can be studied at high resolution in space and time using this programme.28,29
Methanolic extract was found to be the most effective organic extract in comparison to hexane/ethyl‒acetate extract Table 2 and Figure 2. At 10mg/ml concentration, methanolic extract inhibited the growth of S aureus (ZOI: 18.66±0.57), which was nearly two times in comparison to hexane/ethyl‒acetate extract (ZOI: 9.00±0.00). At concentration 1.25mg/ml, methanolic extract still possessed the antibacterial effect (ZOI: 4.00±0.00). Similarly, when pure Quercetin was used to observe its inhibitory potential, at 5 and 2.5mg/ml concentration it showed great potential to inhibit S aureus (ZOI: 25.33±0.57 & 22.66±0.57). As shown in the table, pure Quercetin was very effective even at the concentration of 0.62mg/ml (ZOI: 8.00±0.00). XTT based results also showed the same effectiveness of different organic extracts as revealed by agar‒well assay Table 3. At 20mg/ml concentration, methanolic extract completely inhibited the growth of bacteria and at 10mg/ml concentration, inhibition percentage was dropped to 77.81% as shown in Table 3. Methanolic extract at 5mg/ml of concentration possessed the 56.38% inhibitory power to inhibit the growth of S aureus. Hexane/ethyl‒acetate extract had good inhibitory effect only upto 20mg/ml concentration, where the inhibition percentage was 62.22%. Pure Quercetin showed complete inhibition of bacteria upto 5mg/ml concentration and at concentration of 0.62mg/ml, it still showed effective inhibition (37.66 %). MIC was observed to be 40, 20 and 5mg/ml with Hexane/ethyl‒acetate, methanolic and pure Quercetin extracts, respectively. Whereas, MBC values were found to be 40 and 5mg/ml for methanolic extract and Quercetin, respectively. Biochemical analysis revealed the presence of high concentration of flavonoid and polyphenols and low concentration of coumarin in methanolic extract.
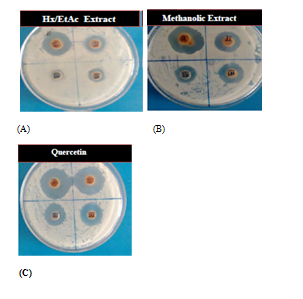
Figure 2 Antibacterial effect of various organic extracts of C officinalis (with concentration 10mg/ml, 5mg/ml, 2.5mg/ml and 1.25mg/ml) against S aureus (A) and (B) antibacterial effect of pure Quercetin (with concentration 5mg/ml, 2.5mg/ml, 1.25mg/ml and 0.62mg/ml) against S aureus (C).
Extracts/Phytochemical |
ZOI (mm±S.D) |
||||
|---|---|---|---|---|---|
10mg/ml |
5mg/ml |
2.5mg/ml |
1.25mg/ml |
0.62mg/ml |
|
Hx/EtAc |
9.00±0.00 |
5.0±0.0 |
3.16±0.28 |
NIL |
‒ |
Methanol |
18.66±0.57 |
12.0±0.0 |
7.33±0.57 |
4.0±0.0 |
‒ |
Quercetin |
‒ |
25.33±0.57 |
22.66±0.57 |
11.16± 0.28 |
8.0±0.0 |
Table 2 Agar‒well diffusion assay results showing zone of inhibition as observed against s aureus
Extract |
OD (mean±SD) |
|||||
|---|---|---|---|---|---|---|
20mg/ml |
10mg/ml |
5mg/ml |
2.5mg/ml |
1.25mg/ml |
0.62mg/ml |
|
Hx/EtAc |
0.349±0.036 |
0.675±0.027 |
0.828±0.040 |
N.E |
N.E |
‒ |
P.I (%) |
(62.22) |
(26.94) |
(10.38) |
‒ |
‒ |
|
Methanol |
C.I |
0.205±0.029 |
0.403±0.042 |
0.571±0.036 |
0.813±0.060 |
‒ |
P.I (%) |
C.I |
(77.81) |
(56.38) |
(38.2) |
(12.01) |
|
Quercetin |
‒ |
C.I |
C.I |
0.116±0.068 |
0.358±0.050 |
0.576±0.034 |
P.I (%) |
C.I |
C.I |
(87.44) |
(61.25) |
(37.66) |
|
Table 3 Measurement of OD (mean ±standard deviation) showing growth of bacteria S aureus in the presence of different concentration of various organic extracts. The well containing antibiotic had OD (0.054±0.086), whereas the well having negative control had the OD (0.924±0.039) CI, Complete Inhibition; NE, No Effect; PI, Percentage Inhibition
All the four enzymes of S aureus are imperative to survival and propagation of bacterial infection. Peptide deformylase (PDF) is a metalloenzyme, which helps in bacterial protein maturation by catalyzing the removal of the N‒formyl group from N‒terminal methionine following the translation process.30 Thus, deformylation plays an important role in bacterial protein maturation. Docking results showed that Alpha cadinol and Quercetin were the most successful phytochemicals to inhibit the PDF enzyme with binding energy of ‒7.7 and ‒7.6kcal/mol, respectively, followed by Scopoletin (‒7.1kcal/mol), and Esculetin (‒7.0kcal/mol) as shown in Table 4 and Figure 3.
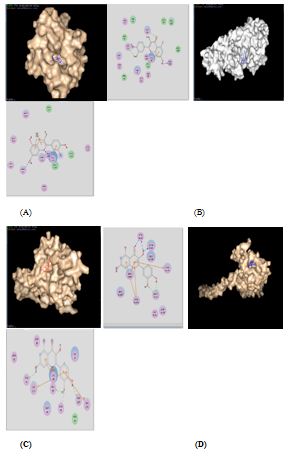
Figure 3 Docking results of various enzymes with quercetin is shown here, as these complexes remains stable during MS simulation study. Each photo is divided into two parts: Left hand portion shows the enzyme‒ligand complex in PYMOL, whereas right hand portion shows the H‒bonding pattern of enzyme‒ligand complex in discovery studio. (A) PDF with quercetin, (B) DNA primase with quercetin, (C) UPPS with quercetin, (D) Gamma hemolysin with quercetin. except gamma hemolysin‒quercetin complex, all the above mentioned complexes are the only stable enzyme‒ligand complex after MD simulation.
|
Gamma hemolysin |
UPPS |
DNA primase |
||
|
|||||
Alpha Cadinol |
‒7.7 |
‒7 |
‒8.3 |
‒6.8 |
|
Scopoletin |
‒7.1 |
‒6.5 |
‒8.1 |
‒7.1 |
|
Quercetin |
‒7.6 |
‒7.4 |
‒5.2 |
‒5.1 |
|
Esculetin |
‒7 |
‒5.9 |
‒8.2 |
‒6 |
Table 4 Free binding energy (kcal/mol) of the various ligands with respective target enzymes
S aureus produced bicomponent pore‒forming toxins (BCPFTs) involving gamma‒hemolysin and leukocidin toxins, which involved in disruption and lysis of erythrocytes and leukocytes. This oligomeric toxin insert a pour into the cell membrane leading to ion influx and efflux reactions, which resulted in initiation of various apoptotic and necrotic pathways, and ultimately cell death.31 As per docking result analysis, this important enzyme was strongly inhibited by Quercetin (‒7.4kcal/mol) and Alpha cadinol (‒7.0kcal/mol) in silico Table 4 and Figure 3. Peptidoglycan is responsible for the mechanical rigidity of cell wall which is required to resist higher osmotic potential and hence maintained cell shape and integrity. Peptidoglycan is the major component of the cell envelope of virtually all bacteria. It has structural roles and acts as a selective sieve for molecules from the outer environment.32 Undecaprenyl Pyrophosphate Synthase is a cis‒prenyltransferase enzyme that catalyzes one of the most important biochemical reactions by successive condensation of eight molecules of isopentenyl pyrophosphate (IPP) with farnesyl pyrophosphate (FPP) to form C55‒undecaprenyl pyrophosphate (UPP). C55‒isoprenol pyrophosphate phosphatase produces Undecaprenyl phosphate (UP) by subsequent removal of the terminal UPP phosphate. Undecaprenyl phosphate involved in transport of various building blocks to allow the synthesis and assembly of peptidoglycans. Peptidoglycan synthesis is, therefore, one of the most important biogenesis pathways in bacteria.32 Alpha cadinol, Esculetin and Scopoletin phytochemicals were found to be the most successful phytochemicals against UPPS enzyme, where Alpha cadinol, Esculetin and Scopoletin showed the excellent binding energy of ‒8.3kcal/mol, ‒8.2kcal/mol and 8.1kcal/mol, respectively Table 4. DNA primases are class of enzymes that synthesize short RNA primers for accurate initiation of DNA replication process. This enzyme is very important for bacterial reproduction and propagation.33 As shown in Table 4 and Figure 3, docking results showed that the DNA primase enzyme was inhibited strongly by Scopoletin (‒7.1kcal/mol), followed by Alpha cadinol (‒6.8kcal/mol), Esculetin (‒6.0kcal/mol) and Quercetin (‒5.1kcal/mol). The PyMOL and Discovery Studio representation of the various stable enzyme‒ligand complexes after computational docking are shown in Figure 3.
Formation of enzyme‒ligand docking complex having good negative energy potential is not enough to proclaim the phytochemical as a good inhibitor. Stability and quality of docked complex is an absolute imperative criteria for a phytochemical to be qualified as a good inhibitor molecule. This can be achieved by molecular dynamics simulation of enzyme‒ligand complex. So, stability and quality of all 16 docked complexes (between 4 enzymes and 4 target proteins, as shown in Table 4 were analyzed by Molecular Dynamics Simulation technique. This technique analyzes the stability factors of enzyme‒ligand complexes which are based on the analysis of RMSD, potential energy and hydrogen energy graphs of the docked complex. The stability of enzyme‒ligand complex is directly proportional to negative potential energy of complex, which further related directly to the inhibitory potential of that respective phytochemical.34‒36 As per analysis of MD simulation results, as shown and explained in Figure 4, it was found that UPPS was strongly inhibited by Quercetin. This exploration is based on the respective P.E (potential energy) graph, the temperature and pressure plot graphs for enzyme‒ligand complex as shown in respective PyMOL window. The active amino acid residue, which formed stable and unbreakable H‒bonds with the Quercetin molecule, was found to be His‒160. RMSD graph of UPPS and Quercetin was found to have stable conformation changes after MD run. The potential energy graph of the UPPS‒Quercetin docking structure shows steady convergence of potential energy after energy minimization step which relaxes the enzyme‒ligand complex structure to ensure that the system has no steric hindrances or inappropriate geometry. In MD simulation, after getting the approximate starting structure, there is a need to equilibrate the solvent and ions around the enzyme‒ligand complex under an NVT/NPT ensemble. As per NVT ensemble plot, it was demonstrated clearly that the temperature of the system quickly reaches the target value (300 K) and remains constant over the remainder of the equilibration. Similarly, NPT plot shows the typical oscillation behavior of the pressure about the preferred average (1.05bar). Numbers of the H‒bond between the enzyme‒ligand complex and their stability was found to be consistent after MD run. Quercetin was again found to be the only stable inhibitor of PDF and DNA primase enzymes as demonstrated by their respective temperature, pressure and potential energy graphs as shown in Figure 5 and Figure 6. Binding affinity of Quercetin towards PDF and DNA primase was found to be highly stable. The active amino acid residue was Met‒177 in case of DNA primase‒Quercetin complex and Gly‒89 in case of PDF‒Quercetin complex. In both cases, H‒bond graph shows that active sites residues have formed stable and proper H‒bonds between the enzyme‒ligand complexes. Similarly, RMSD graph of both enzyme‒ligand complexes were found to be relatively stable with slight structural rearrangement (≤2Ao) throughout the simulation time. Therefore, as per molecular simulation results, the phytochemical Quercetin was found to be the only phytochemical which can be considered as competitive inhibitors against enzymes PDF, UPPS and DNA primase. All the remaining 13pairs of enzyme‒ligand complexes failed during Molecular Dynamics Simulation test, for example, gamma hemolysin was shown to be inhibited by Quercetin as per the docking results Table 4 and Figure 3, but failed to resist the pressure and temperature during simulation which resulted in the breaking of the H‒bond interaction between the enzyme and ligand and the drug moved out of the cavity of the enzyme as shown in Figure 7. At physiological conditions enzyme‒ligand complex, due to dynamic nature, shows structural flexibility. In comparison to docking studies, MD simulations analysis were accredited closer to the physiological conditions and exhibited better binding conformations for docked complex, hence gave almost actual depiction of mode of inhibition.
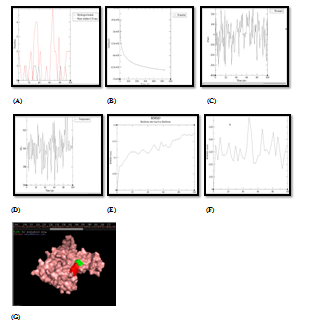
Figure 4 Molecular simulation results of UPPS & quercetin complex. (A) Graph showing hydrogen bonds between the enzyme‒ligand complex within the scale of 0.35nm, (B) Potential energy graph between enzyme‒ligand complex, (C) Pressure graph between enzyme‒ligand complex, (D)Temperature graph between enzyme‒ligand complex, (E) RMSD graph of enzyme, (F) RMSD graph of ligand, (G) Stable enzyme (green colour) & ligand (red colour) complex after MD simulation.
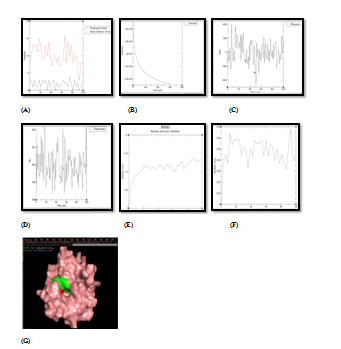
Figure 5 Molecular simulation results of complex PDF and quercetin. (A) Graph showing hydrogen bonds between the enzyme‒ligand complex within the scale of 0.35nm, (B) Potential energy graph between enzyme‒ligand complex, (C) Pressure graph between enzyme‒ligand complex, (D) Temperature graph between enzyme‒ligand complex, (E) RMSD graph of enzyme, (F) RMSD graph of ligand, (G) Stable enzyme (green colour) & ligand (red colour) complex after MD simulation.

Figure 6 Molecular simulation results of complex DNA primase and quercetin. (A) Graph showing hydrogen bonds between the enzyme‒ligand complex within the scale of 0.35nm, (B) Potential energy graph between enzyme‒ligand complex, (C) Pressure graph between enzyme‒ligand complex, (D)Temperature graph between enzyme‒ligand complex, (E) RMSD graph of enzyme, (F) RMSD graph of ligand, (G) Stable enzyme (green colour) & ligand (red colour) complex after MD simulation.

Figure 7 Molecular simulation results of gamma hemolysin and quercetin. (A) Graph showing hydrogen bonds between the enzyme‒ligand complex within the scale of 0.35nm, (B) After simulation, H‒bonding between enzyme‒ligand complex breaks down and the drug molecule moved out from its cavity.
The methanolic extract of the leaves and flowers of C officinalis was found to contain phytochemicals like Quercetin, Scopoletin and Esculetin. Similarly, hexane/ethyl acetate extract contains essential oil which is rich in Alpha cadinol. Methanolic extract showed great inhibitory effect on S aureus, which may be due to many active principles.37,38 Previously, a study demonstrated excellent inhibitory effect of Quercetin against eleven oral pathogenic bacteria like Streptococcus mutans, Streptococcus sanguis, Streptococcus sobrinus and Lactobacillus acidophilu. The antibacterial activity of Quercetin was determined in the form of minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) using agar dilution assay. MIC was found to be in the range of 1‒4mg/ml, whereas, MBC values ranged from 2‒16mg/ml against these oral pathogens.39 In concordance with previous studies, our in vitro study also proved the profound efficiency of methanolic extract of C officinalis and pure Quercetin as of great antibacterial agent against S aureus. As per in silico results Quercetin was found to be the only molecule having strong inhibitory effect against 3 important enzymes of S aureus. It does not means that other phytochemicals like Scopoletin and Esculetin have no contribution in the inhibition of bacterial pathogenesis. They may be involved in different inhibition routes by obstructing other essential biochemical pathways of bacteria. So, as per in silico and in vitro results, Quercetin was found to be the potential antibacterial agent which impedes bacterial growth and pathogenesis by three possible different routes by inhibiting: (i) initiation of replication (inhibiting DNA primase), (ii) Post‒translation modification (inhibiting PDF enzyme) and (iii) bacterial cell wall synthesis (inhibiting UPPS enzyme). Previously, drugs which inhibit PDF enzyme demonstrate potent antibacterial activity against a wide spectrum of bacterial species, including B sublitis, S aureus, E coli, S pneumoniae, S pyogenes, and P aerogenosa.30 No doubt, bacterial growth inhibition is a multifactorial process and inhibition of replication, post‒translational modification and cell wall precursor’s enzyme could be some of the pathways inhibited by Quercetin to control bacterial pathogenesis. However, further studies under in vitro conditions are necessary to authenticate the potential of this phytochemical as an effective drug against S aureus and other pathogenic bacteria.
Plants derived medicines are relatively economical to synthetic drugs. So, it appears prudent to evaluate antibacterial potential of unexplored medicinal plants. The last three decades witnessed a tremendous growth in the chemical and pharmaceutical studies of medicinal plants for isolation of novel active principles having antibacterial, antiviral and anticancerous potential. But, the information regarding the mechanism of inhibition of these novel phytochemicals remains both scanty and vague. In our study, we have successfully demonstrated that Quercetin, which is found in methanolic extract of C officinalis has great potential to inhibit the growth of S aureus by inhibiting 3 essential enzymes of replication, post‒translational and cell‒wall synthesis pathways. The in vitro results showed that 20mg/ml concentration of the methanolic extract was found to completely inhibit the bacterial growth. Virtual screening like docking and simulation are fast and cheap methods that give sufficient primary information of inhibition pathway of phytochemicals against essential enzymes of various pathogens.40 Further, in silico structure based drug designing can be used to improve the efficacy of inhibitory effects of Quercetin like natural molecules. This study unbolts the opportunity for further research on these types of natural compounds, as it is evident from the facts that these compounds have great potential to be used as lead molecules in future development of drugs.
Authors are grateful to Dr. Vinod Mittal (M.D), Dolphin (P.G) College of Science and Agriculture for providing financial supports to this study. We are highly indebted to the Principal, Dr. S.P. Jindal, for his encouragement to present research and giving final shape to this paper.
Author declares there is no conflict of interest.

©2018 Pahal, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.