MOJ
eISSN: 2374-6912


Research Article Volume 3 Issue 2
1Department of Otolaryngology, Head and Neck Surgery, The First Affiliated Hospital of Anhui Medical University, China
2National Laboratory of Molecular Oncology, Chinese Academy of Medical Sciences and Peking Union Medical College, China
3Department of Otolaryngology, Head and Neck Surgery, The Chinese PLA General Hospital, China
4Department of Automation, Tsinghua University, China
Correspondence: Liyong Zhang, MOE Key Laboratory of Bioinformatics, Tsinghua University, Beijing 100084, China
Co-correspondence: Mingbo Liu, Department of Otolaryngology, Head and Neck Surgery, The Chinese PLA General Hospital, Beijing, 100853,China
Received: February 08, 2016 | Published: March 3, 2016
Citation: Liu Y, Liu Z, Liu M, et al. Profiling of MicroRNA expression in head and neck cancer. MOJ Cell Sci Rep. 2016;3(2):53-58. DOI: 10.15406/mojcsr.2016.03.00053
MicroRNAs (miRNAs) are a class of small non-coding RNAs that play regulatory roles by repressing translation or cleaving RNA transcripts. Accumulating evidence suggests that miRNAs are aberrantly expressed in many human cancers and that they play significant roles in the initiation, development and metastasis of human cancers. Genome-wide miRNA expression profiling provides information on the aberrant expression of miRNAs in cancers rapidly and precisely. Many human cancers have been profiled but there are few published studies in head and neck cancer. In this study, the expression of miRNAs was determined in five patient-matched head and neck tissues using miRNA microarray. Expression of eight miRNAs including miR-1, miR-126* (*, refer to miR-126-3p), miR-139, miR-375, miR-181a, miR-181b, miR-21 and miR-7 were validated using real-time RT-PCR and/or Northern blot analysis. This study provides the largest genome-wide survey of mature miRNA transcripts in head and neck cancer. The implication of aberrantly expressed miRNAs in head and neck cancer suggested their involvement in the development and progression of these malignancies. Further experiments of these deregulated miRNAs will help elucidate their roles in tumorigenesis and furnish their potential clinical usage for early diagnosis, prognosis and therapy of head and neck cancer.
Keywords: miRNA, head and neck cancer, cancer genetics
HN, head and neck; HPV, human papilloma virus; SCC, squamous cell carcinomas; LCM, laser capture microdissection; PGR, photo generated reagent; FAK, focal adhesion kinase
Head and neck (HN) cancer is the sixth most common cancer worldwide, with approximately 650,000 new cases reported annually.1-3 In the United States, there were 49,000 new cases in 2010, with 11,000 death.3,4 Aggravating risk factors include tobacco smoking, alcohol consumption, betel chewing, and human papilloma virus (HPV) infection.3,5 Head and neck cancer usually originates in the squamous cells lining the moist, mucosal surfaces inside the head and neck, such as inside the mouth, the nose, and the throat. All these squamous cell cancers are referred to as squamous cell carcinomas (SCC) of the head and neck. In spite of the overall incidence decrease of HNSCC in the United States during the past thirty years, there is a dramatic incidence increase for squamous cell malignancies of the base of tongue, and tonsil, especially in the populations who are young-to-middle-age patients. This is most likely coming from the rising incidence of HPV-associated HNSCC.6 Although treatments of head and neck cancer include surgery, radiation therapy, chemotherapy, targeted therapy, or a combination of treatments, the 5-year survival rate for locoregionally advanced head and neck cancer patients is still poor.3,7-9 The genetic alteration of cells in wide preneoplastic fields results in locoregional recurrence and second primary cancer. Half of all individuals still die from their disease. The characterization of the mechanisms involved in tumorigenesis and the identification of potential markers allow the identification of patients with biologically aggressive tumors, which are of great interest for the effective management of HNSCC patients.
MicroRNAs (miRNAs) are short non-coding RNAs identifying in plants, animals, and some viruses working in RNA silencing and post-transcriptional regulation of gene expression. While approximately 20% of human miRNAs are transcribed by RNA polymerase III, most of the miRNAs are transcribed by the RNA polymerase II to produce a primary-microRNA (pri-miRNA), which is then processed as ~60-nt long pre-miRNA hairpin intermediates. The intermediates are further processed to ~22-nt miRNA duplexes to negatively regulate protein expression by mRNA degradation or protein translation repression through base-pairing to complementary sites on the targets.10 Mounting evidences have indicated the pivotal role of miRNAs in many biological processes including cell motility, cell proliferation, apoptosis, cell metabolism, cell differentiation, morphogenesis, and developmental timing.11,12 Numerous studies have revealed the importance of miRNAs in carcinogenesis13,14 and defined that high-throughput miRNA profiling is an informative tool to determine the lineage of development and the status of differentiation in various human malignancies. However, very little attention has been paid to head and neck cancer in genome miRNA analysis. Urgent priorities are needed to identify novel gene(s) or pathways that may elucidate the mechanisms of this disease. In this study, miRNA microarray was utilized to measure the relative expression of miRNAs in head and neck cancer. In addition, we attempted to seek associations in the literature between these miRNAs and their potential targets for a role in carcinogenesis of head and neck.
Patients and samples
The tissue samples (Table 1) were obtained from patients with head and neck cancer diagnosed by the pathologists in the First Affiliated Hospital of Anhui Medical University with informed consent and agreement. All specimens were snap frozen in liquid nitrogen immediately after surgical resection, and stored in -80℃ until used. The study was approved by the medical ethics committee of Tsinghua University.
To obtain pure cancer and/or normal cells, laser capture microdissection (LCM) was carried out by the qualified pathologist. Total RNAs were extracted by using the mirVanaTM miRNA Isolation Kit (Ambion, Austin, Texas) according to the instructions of the manufacturer. The RNA quality was evaluated by 15% denaturing polyacrylamide gel electrophoresis and spectrophotometry (Eppendorf BioPhotometer, Eppendorf, Hamburg, Germany).
Sample No. |
Usage |
HN00051 |
LCM, qRT-PCR |
HN00143 |
LCM, qRT-PCR |
HN00165 |
LCM, qRT-PCR |
HN00174 |
LCM, qRT-PCR |
HN00197 |
LCM, qRT-PCR |
HN00212 |
LCM, qRT-PCR |
HN00214 |
LCM, qRT-PCR |
HN00489 |
LCM, qRT-PCR |
HN00542 |
LCM, qRT-PCR |
HN00554 |
LCM, qRT-PCR |
HN00555 |
LCM, qRT-PCR |
HN00556 |
LCM, qRT-PCR |
HN00571 |
LCM, qRT-PCR |
HN00581 |
LCM, qRT-PCR |
HN00587 |
miRNA microarray, Northern blot, LCM, qRT-PCR |
HN00596 |
LCM, qRT-PCR |
HN00607 |
LCM, qRT-PCR |
HN00647 |
miRNA microarray, Northern blot, LCM, qRT-PCR |
HN00653 |
miRNA microarray, Northern blot, LCM, qRT-PCR |
HN00654 |
LCM, qRT-PCR |
HN00719 |
miRNA microarray, Northern blot, LCM, qRT-PCR |
HN00728 |
miRNA microarray, Northern blot, LCM, qRT-PCR |
Table 1 Summary of human head and neck cancer tissues used in this study
µParaflo™ MicroRNA microarray Assay
High-throughput miRNA gene expression analysis for five snap-frozen patient-matched head and neck samples (HN00587T/C, HN00647T/C, HN00653T/C, HN00719T/C and HN00728T/C) was executed by LC Sciences (http://www.lcsciences.com/; Houston, TX. The assay started from 2 to 5µg total RNA sample, which was size-fractionated using a YM-100 Microcon centrifugal filter, and the small RNAs (<300nt) isolated were 3’-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye staining; two different tags were used for the two RNA samples in dual-sample experiments. Hybridization was performed overnight on a µParaflo microfluidic chip using a micro-circulation pump (Atactic Technologies). On the microfluidic chip, each detection probe consisted of a chemically modified nucleotide coding segment complementary to target microRNA from miRBase (http://microrna.sanger.ac.uk/sequences/) or other RNA (control or customer defined sequences) and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. The detection probes were made by in situ synthesis using PGR (photogenerated reagent) chemistry. The hybridization melting temperatures were balanced by chemical modifications of the detection probes. Hybridization used 100µl 6xSSPE buffer (0.90M NaCl, 60mM Na2HPO4, 6mM EDTA, pH6.8) containing 25% formamide at 34°C. After hybridization detection used fluorescence labeling using tag-specific Cy3 and Cy5 dyes. Hybridization images were collected using a laser scanner GenePix 4000B (Molecular Device, Sunnyvale, CA) and digitized using Array-Pro image analysis software (Media Cybernetics). Numerical intensities were extracted for control, background, and miRNA probes and converted into Microsoft Excel spreadsheets.
Data were analyzed by first subtracting the background and then normalizing the signals using a LOWESS filter (Locally-weighted Regression). For two color experiments, the ratio of the two sets of detected signals (log2 transformed, balanced) and p-values of the t-test were calculated; differentially detected signals were those with less than 0.01 p-values. The TIGR MeV (Multiple Experimental Viewer) (the Institute for Genomic Research) microarray program was used for clustering analysis.
Real-time RT-PCR
Total RNAs were isolated using mirVanaTM qRT-PCR miRNA Detection Kit (Ambion, Austin, Texas) according to the instructions of the manufacturer. Twenty ng of total RNAs were used for reverse transcription of cDNA. The PCR reaction was performed in iCycler (Bio-Rad, Hercules, CA) consisting of appropriate number of cycles (95°C for 15 s, 60°C for 30s) after an initial denaturation step (95°C for 3min) using mirVana™ qRT-PCR Primer Sets (Ambion, Austin, Texas). In addition, the PCR products were separated on 3.5% agarose gel and visualized with ethidium bromide on the ChemiImager Imaging System 5500 (Alpha Innotech, San Leandro, CA). The 5 S RNA was used as an internal control.
Northern blotting
Northern blotting was performed as previously described.15 Briefly, 10µg of total RNAs from each sample was fractionated on 15% denatured polyacrylamide gel containing 8M urea, then transferred onto Bright Star®-Plus positively charged nylon membrane (Ambion, Austin, Texas) using semi-dry transfer apparatus (Bio-Rad, Hercules, CA). Blots were pre-hybridized at 65°C for 1hour in pre-hybridization buffer (200mM Na2HPO4, pH7.0, 5% SDS) and subjected to hybridization with 32P-labeled miR-181a, or -21 probes overnight. 32P-labeled U6 probe was used as a loading control. Following hybridization, membranes were washed twice for 10 min each at 25°C using low-stringency buffer (25mM Na2HPO4, pH7.5, 5% SDS, 3×SSC), and once for 10 min at 42°C using high-stringency buffer (1% SDS, 1×SSC). The blots were exposed to X-ray film (PIERCE, Rockford, IL) in -80°C freezer. The oligonucleotides used as probes are the complementary sequences of the mature miRNA (miR Registry, http://www.sanger.ac.uk/Software/Rfam/mirna/): miR-181a: 5’-actcaccgacagcgttgaatgtt-3’; and miR-21: 5’-tcatcttcagtctgataagcta-3’. An oligonucleotide complementary to the U6 RNA (5’-gcaggggccatgctaatcttctctgtatcg-3’) was used to normalize expression levels.
Altered expression of miRNAs in head and neck cancer
Total RNAs from 5 pairs of snap-frozen patient-matched head and neck samples were sent to LC Sciences (Houston, TX) for miRNA microarray assay. The miRNA expression profiles were defined between tumor (designated as “T”) and adjacent normal tissues (designated as “C”) after performing the correlation analysis. By executing the correlation analysis from 2 chips, the system related biases coming from labeling, handling can be eliminated and therefore calls can be narrowed down to the true biological differences. Differentially expressed miRNAs (at P < 0.01) between tumor and adjacent normal tissues were identified. Table 2 showed the eight miRNA candidates chosen for further validation. Figure 1 showed the unsupervised hierarchical clustering of miRNA expression.
The 5 paired samples are in columns and the miRNAs are in rows. T, tumor tissue; C, adjacent normal tissue.
signal intensity miRNA candidates |
HNSFT00587 |
HNSFT00647 |
HNSFT00653 |
HNSFT00719 |
HNSFT00728 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
Tumor |
Normal |
Tumor |
Normal |
Tumor |
Normal |
Tumor |
Normal |
Tumor |
Normal |
|
miR-21 |
70553.63 |
19652.55 |
40117.68 |
2568.90 |
67534.99 |
21435.48 |
73021.53 |
12934.91 |
8230.68 |
8099.10 |
miR-7 |
735.92 |
222.63 |
_ |
_ |
1691.31 |
136.20 |
541.58 |
12.60 |
109.68 |
5.24 |
miR-181a |
4170.48 |
1736.07 |
_ |
_ |
4057.30 |
1883.81 |
2119.26 |
763.45 |
2961.16 |
1319.05 |
miR-181b |
3200.38 |
1088.88 |
1356.68 |
355.22 |
3030.89 |
1293.69 |
1603.81 |
584.52 |
3012.02 |
1166.77 |
miR-1 |
30.8 |
3458.69 |
4.88 |
577.67 |
58.15 |
1299.80 |
258.39 |
31317.54 |
2421.02 |
24670.59 |
miR-126* |
92.78 |
3570.01 |
_ |
_ |
94.76 |
4008.85 |
475.66 |
3157.70 |
61.29 |
796.16 |
miR-139 |
7.38 |
270.99 |
_ |
_ |
18.64 |
425.76 |
28.35 |
171.23 |
11.22 |
313.75 |
miR-375 |
476.46 |
7529.95 |
9.14 |
356.57 |
286.86 |
5899.93 |
109.30 |
626.28 |
128.59 |
16200.45 |
Table 2 Eight miRNAs of interest chosen from the differentially expressed transcripts for validation
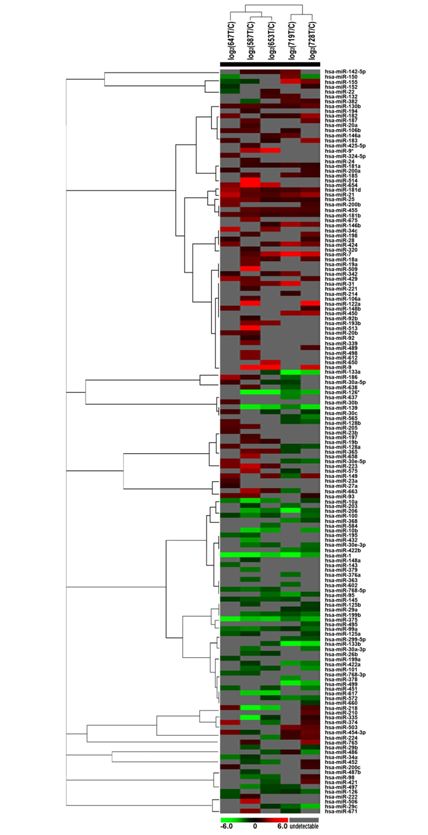
Figure 1 Unsupervised hierarchical clustering of miRNA profiles of 5 paired samples from head and neck cancer using Cluster 3.0.
Validation of the miRNA microarray
Eight differentially expressed miRNA candidates miR-1, -126* (-126-3p), -139, -375, -181a, -181b, -21 and -7 were chosen to confirm the miRNA microarray results by Northern blotting and/or real-time PCR. The results indicated that comparing their adjacent normal counterparts, miR-181a, -181b, -21, and -7 were upregulated in cancer, while miR-1, -126* (-126-3p), -139, and -375 were down-regulated in cancer (Figure 2). These results were consistent with the results from miRNA microarray assay.
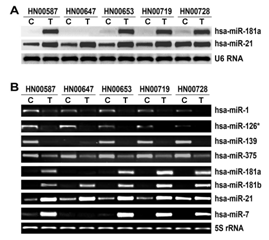
Figure 2 Validation of the miRNA microarray results.
Expression of miRNA candidates in laser capture micro dissected (LCM) samples
Laser capture microdissection was performed for twenty-one patient-matched head and neck samples, and the total RNAs were extracted for real-time PCR. The results showed that, in comparison with their adjacent normal counterparts, miR-181a, -181b, -21, and -7 were increased in cancer cells while miR-1, -126* (-126-3p), -139, and -375 were decreased in cancer cells (Figure 3).
Real-time RT-PCR of miR-1, -126* (-126-3p), -139, -375, -181a, -181b, -21 and -7 in patient-matched laser capture microdissected head and neck samples. The 5S rRNA was used as an internal control. T, tumor cells; C, adjacent normal cells.
Carcinogenesis is a complicated and multi-step process in which many genes are implicated.16 MicroRNAs constitute a novel class of gene regulators that are ~22 nt small non-coding, single-stranded RNAs.17 It has been found that they are abnormally expressed in diverse cancers.18-20 And studies have shown that some miRNA signatures could be correlated well for the classifications with different clinicopathologic parameters.21,22 To develop rational approaches for the diagnosis and treatment of cancer depends on identifying new biomarkers and understanding the molecular mechanisms that underlie tumor formation and progression. In this aspect, studies that seek to identify dysregulated miRNAs in neoplasm are critical. Thus, research efforts aimed at systematically identifying the miRNA expression profiles between normal and tumor cells are critically needed. In this study, we have defined the miRNA expression profiles in 5 patient-matched human head and neck samples using miRNA microarray. After data analysis, we obtained the differentially expressed miRNAs (P<0.01) between normal and cancer samples. The profiles of miRNAs from samples HN00587, HN00653, HN00719 and HN00728 could be clustered while miRNAs from sample HN00647 couldn’t be fitted in. This might be caused by the exact location of the tumor, the stage of the cancer, or the patient’s age and general health, such as infection with cancer-causing types of human papillomavirus (HPV). Some miRNAs were significantly changed in their expression level between neoplastic tissues and normal ones. Among them, four miRNA candidates (miR-181a, miR-181b, miR-21, and miR-7) showed dramatic upregulation in tumor tissues, while other four miRNAs were downregulated compared with the patient-matched normal controls, including miR-1, miR-126* (miR-126-3p), miR-139, and miR-375. To confirm the results of the microarray analysis we performed northern blot and/or real-time PCR on a limited number of samples. Northern blot and/or qRT-PCR confirmed the results obtained by microarray analysis.
Recent advances in the understanding of the oncogenesis have revealed multiple deregulated miRNAs.23,24 Most of the differentially expressed miRNAs in head and neck cancer we found showed are consistent with previous studies.25 For example, miR-1, a highly conserved miRNA in the musculatures of flies, mice and humans and a well characterized as muscle-specific miRNA, which was downregulated in head and neck cancer from this study, was consistently down-regulated in human prostate tumors.26-28 In colorectal cancer, miR-1 directly decreased MET oncogene both at RNA and protein level, and its recovery resulted in reduction of cell proliferation and motility driven by MET, identifying that miR-1 served as a tumor suppressor.29 In gastric cancer, miR-1 inhibited cell proliferation and migration by targeting MET.30,31 MiR-126* (miR-126-3p), which was downregulated in head and neck cancer from this study, changed in the expression levels in various cancer cells originating from breast, lung, cervix, bladder, prostate, colon, liver, prostate, esophageal, or gastric cancer, as well as leukemia.32 It repressed recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit metastasis in breast cancer,33 and suppressed tumor metastasis and angiogenesis of hepatocellular carcinoma by targeting LRP6 and PIK3R2.34 MiR-21, which was upregulated in head and neck cancer from this study, has been shown to be upregulated in different cancers including pancreatic neuroendocrine tumors, and might play an important role in preventing apoptosis.23,24 Moreover, it was overexpressed in pancreatic cancer and its increase predicted limited survival in patients with node-negative disease.35 MiR-375, an islet-specific miRNA to regulate insulin secretion,36 which was downregulated in the present this study, was also down-modulated in pancreatic cancer.36 MiR-139, downregulated in head and neck cancer, targeted the type I insulin-like growth factor receptor to inhibit invasion and metastasis in colorectal cancer,37 and decreased Rho-Kinase 2 to suppress the metastasis and progression of hepatocellular carcinoma.38 More and more evidences implicated miR-7 in numerous signaling cascades and diseases. miR-7 directly regulated α-synuclein in Parkinson disease.39 In pancreatic β-cells, the inhibition of miR-7 induced mTOR signaling, which subsequently stimulated cellular proliferation.40 This proposes miR-7 as a putative cause of low β-cell renewal and a possible therapeutic target in diabetes. miR-7 also played a tumor-suppressive role by directly targeting and decreasing central oncogenic factors in cancer-associated signaling pathways.41 In consistency with such a role for miR-7, it was observed to be the most reduced miRNA in cancer stem-like cells.42 The tumor-suppressive role of miR-7 could be elucidated in part by modulating Kruppel-like factor 4 (KLF4) to inhibit brain metastasis in breast cancer.42 In addition, miR-7 upregulated E-cadherin indirectly through insulin-like growth factor 1 receptor42 and focal adhesion kinase (FAK),41 which led to reduced epithelial-to-mesenchymal (EMT) transition, reduced anchorage-independent growth and suppression of metastasis. In colorectal cancer, miR-7 was one of the most decreased miRNAs and it was shown to decrease p53 by targeting the oncogenic YY1 transcription factor.41 Although mounting evidences supported that miR-7 was a tumor-suppressor, the opposite effects have also been documented. In lung carcinomas, poor prognosis was found to be associated with miR-7 overexpression through a Ras/ERK/Myc pathway.43 While miR-181a and miR-181b, increased in head and neck tumor in this study, were found decreased in prostate cancer and glioblastoma,20,44 suggesting the same microRNA could exert opposite effects in different organs.
Genetic changes are the key elements to the development of cancer. And these genetic changes endow the cancer cells with many of the hallmarks of cancer, such as self-sufficient growth and resistance to anti-growth and pro-death signals.3 Although mounting evidences have shown that genetic changes occur within cancer cells themselves, such as activated oncogenes or malfunctioned tumor suppressors, are responsible for many aspects of cancer development, it is likely that tumor progression is through multiple mechanisms that are not yet fully understood. One possible mechanism is that tumor promotion and progression are dependent on physiologic responses provided by supportive tissues of the tumor environment but that are not necessarily cancerous themselves. To verify whether the dysregulation of miRNAs was effectively restricted to the tumor cells in the samples, laser capture microdissection was performed to obtain pure cancer cells and normal cells for real-time PCR. As shown in Figure 3, real-time PCR results also confirmed the dysregulation of miRNAs cancer samples compared to normal samples. So far this is one of the first studies of this nature in head and neck cancer tissues.
In summary, the goal of this study was to profile the relative expression of miRNAs in head and neck cancer samples. How the miRNAs of interest play in cancer remains mostly unknown, and their clinical utility needs to be determined. However, these findings provide a base foundation for further characterization of miRNAs functions in head and neck cancer.
None.
The author declares no conflict of interest.

©2016 Liu, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.