MOJ
eISSN: 2374-6912


Stem cells give rise to almost 200 different cell types that are present in a mammalian organism. In spite of the fact that these distinct cell types have different functions and morphologies, they descend from a common ancestor cell and essentially share the same DNA. The underlying cause for the rise of specific cell types is not genetic differences; it is mostly due to how the genetic information is interpreted. The epigenome, which consists of several degrees of regulatory mechanisms, dictates the gene expression profile of a certain type of cell. Eukaryotic DNA is packaged into chromatin, folded and compacted which in turn affects its functionality as certain regions of DNA will not be accessible whereas some other regions will be easier to access for effect or proteins and modifiers to bind. The chromatin dynamics are modulated by several machineries including but not limited to histone PTMs and their variants, DNA methylation and RNA interference, which have crucial roles in regulating stem cell differentiation, cell fate determination and lineage specification.
Keywords: stem cells, epigenetics, chromatin, histone modifications, DNA methylation, RNAi
Pluripotent stem cells have the capability of differentiating into cells from all three germ layers endoderm, mesoderm and ectoderm; as well as an unlimited potency to self-renew, maintaining their stem cell identity. Embryonic stem cells that are derived from the inner cell mass of the mammalian blastocyst can give rise to a fully developed viable embryo with more than 200 different cell types, which essentially share the same genetic information, namely the DNA. The establishment of distinct cellular identities requires differential interpretation of this genetic information in the form of differential gene expression patterns, which is largely influenced by DNA accessibility and is modulated by mechanisms that are beyond DNA sequence, indicating epigenetic regulation. In eukaryotes, DNA is compacted into a macromolecular structure called chromatin. The packaging of DNA into chromatin is done in a dynamic manner so that the DNA can still be accessible to carry out cellular functions such as replication, transcription and DNA repair. The naked DNA first gets wrapped around the histone octamer containing two copies of each core histone H2A, H2B, H3 and H4, which constitutes the first step of compaction called the “beads on a string” conformation.1 Next, the linker histone H1 binds to the nucleosome at the DNA entry-exit point and further condenses the DNA into 30nm chromatin fiber. Metaphase chromosomes represent the most compact form of chromatin2
Chromosomes are comprised of two structurally and functionally distinct domains; euchromatin represents the transcriptionally active, loosely packaged and gene-rich regions whereas heterochromatin is highly condensed and gene-poor.3 There are also two types of heterochromatin; constitutive heterochromatin is found mainly at the centromeres, telomeres and repeats, safeguarding genome integrity, and is poorly expressed whereas facultative heterochromatin can be converted to euchromatin depending on the cell type and developmental stage at specific loci.4 The transition between different chromatin forms at different genomic loci leads to the establishment of distinct gene expression patterns for each cell type. Chromatin in stem cells is lightly packed and the actively transcribed genes are mainly the ones that are required for maintaining pluripotency. However, as they gradually lose their pluripotent characteristics and start differentiating, pluripotency genes are turned off and lineage specific gene expression takes over. Previous studies indicate a close connection between stem cell differentiation and widespread epigenome remodeling.5 Furthermore, the epigenetic landscape set during differentiation is important in maintenance of the cell fate and lineage commitment, acting as a cellular memory.6
Histone modifications and variants
Histones are major players of chromatin dynamics and transcriptional activity. Specific histone post-translational modifications (PTMs) and incorporation of certain histone variants are associated with euchromatin and active transcription while others might be found at heterochromatic regions and repress gene expression.7–9 Histones are small basic proteins that are rich in arginine and lysine amino acids. The tripartite structure of histones that are composed of a globular domain and unstructured N- or C-terminal tails allow these amino acids and also others to be subjected to several covalent modifications such as methylation, acetylation, phosphorylation.10 The list of histone PTMs has significantly grown in recent years as many new sites of modifications and their modifiers have identified.11 The “histone code hypothesis” suggests that distributions of histone PTMs form a signature that is indicative of the chromatin state of a given loci.12,13 For instance, euchromatin is generally associated with high levels of histone acetylation such as H3K9ac and H4K16ac which are found at the promoters of actively transcribed genes.14,15 while heterochromatin is enriched in repressive marks such as H3K9me3 and H3K27me3. The gene expression regulation during stem cell differentiation into lineage specific cells requires action of histone modifying enzymes. For instance, removal of certain histone acetylation marks by histone deacetylases is crucial for neuronal precursor cells to undergo neurogenesis.16 The action of histone modifications and their enzymes is often accompanied by other chromatin modifiers and DNA binding proteins as well. Studies indicate that increased levels of repressive histone marks such as H3K27me3 elevate levels of the polycomb group of protein complex (PcG)-associated BMI-1 (B-lymphoma Mo-MLV insertion region-1 homolog), which results in decreased neural stem cell proliferation.17,18
DNA methylation
Heterochromatin usually has high levels of DNA methylation.4 In mammals, DNA is methylated at the CpG dinucleotides by DNMT3A and DNMT3B during embryonic development and inherited to the next generation in a semi-conservative manner in every cell division.19 DNMT1 methylates the newly synthesized, unmethylated strand to ensure the maintenance of the methylated state.20 As DNA methylation (DNA-me) is closely linked with gene expression regulation, it plays a major role in stem cell differentiation and lineage specification. The stem cell genome is highly hypomethylated whereas the global methylation levels generally increase as cells commit to a certain fate and differentiate.21 The expressions of the driver genes of pluripotency; Oct4, Nanog and Sox2 are largely regulated by promoter methylation, as increasing DNA-me levels results in their downregulation and loss of pluripotency.22–24 Studies indicate that neural stem cells express DNMTs during the course of cellular specification and differentiation.25 Likewise, reduction in DNA methylation levels by removal of Dnmt1 causes intestinal epithelium cells to differentiate less.26 Studies also indicate that stem cells start randomly differentiating when they are treated with chemicals that result in DNA demethylation.21
RNA-based mechanisms
Non-coding RNAs have been linked with initiation of heterochromatin formation and transcriptional silencing. For instance, the long non-coding RNA Xist (X-inactive specific transcript) plays a crucial role in X chromosome inactivation.27 Likewise, non-coding RNA mediated targeting of heterochromatinization is widely studied.28–30 There are also small RNA molecules comprising of 20-30nt such as micro-RNAs (miRNAs), small interfering-RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs) that function in modulating gene expression. These RNA based mechanisms that are collectively referred to as RNA-interference (RNAi) are involved in gene silencing at the post-transcriptional level. The expression patterns of small RNAs change among different tissues at different developmental time points; which in turn affects the expression levels of their downstream target genes. Hence, the RNA molecules within the RNAi machinery play a major role in tissue-specific gene expression during development.31 In line with this, there are several studies in the literature pointing towards major roles for miRNAs in differentiation of stem cells into neuronal cells.17 For instance, miRNA-124 is specifically expressed in the brain and it is one of the major players that regulate the rate of neurogenesis.32 Another miRNA, namely Let-7 is also associated with neuronal cell differentiation through transcriptional repression (Figure 1).20
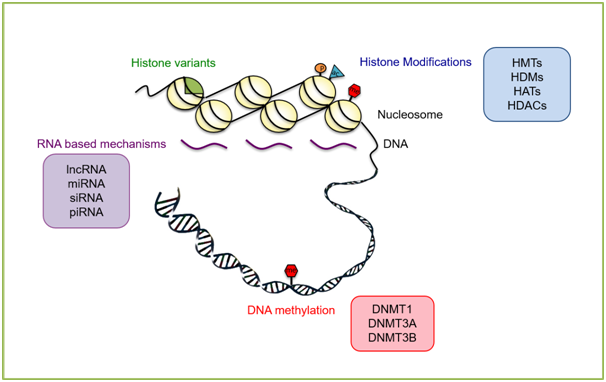
Figure 1 Selected epigenetic regulation mechanisms that modulate chromatin dynamics during stem cell differentiation. Histone modifications set by histone methyltransferases (HMTs) and histone acetylases (HATs) and removed by histone demethylases (HDMs) and histone deacetylases (HDACs), incorporation of certain histone variants, RNA based mechanisms and DNA methylationare key players in setting lineage-specific gene expression profiles. See text for more detail.
Stem cells have opened a very promising avenue for novel therapeutic approaches. Although they were first discovered 40years ago, stem cell research has attracted great attention within the past 20years. The establishment of embryonic stem cell cultures in vitro, cloning animals from stem cells and the identification of cancer stem cells enabled groundbreaking developments in the field. Despite the fact that stem cell therapy could be the ultimate answer for most diseases, the difficulty in finding and isolating them from adults limits its applications. However, with the discovery of induced pluripotent stem cells, it is now possible to de-differentiate lineage specific cells into stem cells, which allows trans-differentiation to generate desired cell types required for treatment. Induction of stem cells from patient’s own somatic cells also means lower risk of rejection due to incompatibilities after transplantation. All these mechanisms of stem cell differentiation de-differentiation of somatic cells and trans-differentiation with the aim of personalized stem cell therapy requires a complete view of epigenetic machineries involved in these processes. Therefore, future studies examining the details of histone PTMs and their variants, DNA methylation and RNA interference in relation to stem cell research is necessary.
The author declares that there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.