MOJ
eISSN: 2374-6912


In this study, extended spectrum beta-lactamase (ESBL) genes among Acinetobacter baumannii isolated from hospitalized patients with burn infections were evaluated. From December 2016 to March 2017, sixty-five isolates (72.2%) of A. baumannii were obtained from 90 burn samples of patients which were hospitalized in two hospitals in Baghdad, Iraq. The identification of A. baumannii strains was conducted by API 20NE and the presence of blaOXA-51 gene. Kirby-Bauer disc diffusion method was used for antibiotic susceptibility test. The frequency of ESBL producers was evaluated by double disk synergy test (DDST) and prevalence of blaTEM, blaSHV, blaCTX-M genes were assayed by PCR. The results revealed that blaOXA-51 gene was found in all A. baumannii isolates as a species-specific gene. Of sixty-five isolates, 40 (61.53%) A. baumannii were ESBL producers according to DDST. High rates of resistance to cefotaxime, ceftazidime, piperacillin, and cefepime antibiotics were detected among ESBL producing A. baumannii isolates. The rate of resistance to other antibiotics was as the follows: 29(72.5%) to meropenem, 32(80%) to imipenem, 30 (75%) to piperacillin/tazobactam, 26 (65%) to ampicillin/sulbactam. The prevalence of blaTEM, blaCTX-M, blaSHV genes among ESBL producing A. baumannii isolates was 30 (75%), 18 (45%) and 11 (27.5%) respectively. Our results demonstrated the role of blaTEM gene in the resistance of beta-lactam antibiotics in the local strains of burn infections. The high rate of resistance in ESBL producing A. baumannii strains detected in this study highlights the necessity for the quick detection of ESBL isolates and the antimicrobial management, particularly in burn units.
Keywords: A. baumannii, ESBL genes, burns
Acinetobacter baumannii is an important cause of nosocomial infections and a leading cause of mortality and morbidity among hospitalized patients and has been associated with a wide variety of diseases in the intensive care units (ICU). Widespread dissemination of A. baumannii infections, which are resistant to beta-lactams antibiotics particularly to the 3rd generation of cephalosporins and carbapenems, has become a globally significant problem.2 These extended-spectrum β-lactamases (ESBLs) are enzymes, plasmid mediated and produced by gram negative bacilli that mediate a resistance to penicillins, cephalosporins, and monobactams.3 These ESBLs are commonly recognized in Enterobacteriaceae, Pseudomonas aeruginosa, and A. baumannii and are found worldwide.4 Extended-spectrum ß-lactamases (ESBLs) of A. baumannii are encoded by TEM-type, SHV-type and CTX-M-type genes which are subjected to phenotypical resistance to penicillins and 3rd generation cephalosporins. These enzymes are mostly plsmid mediated and most are parts of the TEM and SHV families.2,5 Different microorganisms are related to contaminations in hospital environments, but the main pathogens include oxacillin-resistant Staphylococcus aureus (ORSA), vancomycin-resistant Enterococcus sp. (VRE), extended-spectrum beta-lactamases (ESBL), and carbapenem-resistant Acinetobacter baumannii.6,7 A. baumannii is able to survive on human skin, equipment, and objects in different sections, especially burn centers and ICUs for a long time.8 The major problem of concern with A. baumannii is the choice of suitable antibiotic for treatment, especially in nosocomial infections.9 The aim of this study was to investigate the detection and distribution of ESBLs encoding genes and its drug resistance against carbapenems and cephalosporins among A. baumannii strains isolated from patients with burn infections.
Sampling and isolation of bacteria
The study was initiated in December 2016 by collecting 90 samples from burn infections of burn units in 2 hospitals of Baghdad City in Iraq. The samples were collected by sterile swabs which used moistened with Trypticase Soy Broth (TSB) medium. [Based on what type of test characterization and identification of isolates were performed?] Immediately after the collection, they were again stored in the medium and incubated at 37oC for 24 hours. After the growth in TSB, the samples were cultured on Blood agar and MacConkey agar incubated at 37oC for 24 hours. The identification of the isolates was accomplished by API 20NE and confirmed by tracking the blaOXA-51-like carbapenemase gene, which is intrinsic to this species, using single PCR.10
Antibiotic susceptibility testing
The susceptibility of A. baumannii isolates to carbapenems and cephalosporins (Mast, UK) was tested by Kirby-Bauer disk-diffusion method. The antibiogram procedure was performed as the manufacturer which manufacture constructed. In brief, 1.5*108 CFU/ml of bacterial suspension, equivalent to McFarland Turbidity Standard No. 0.5, was transferred on Muller-Hinton agar medium (Merck, Germany) and antibiogram disks containing: cefipime (FEP:30μg), cefotaxime (CTX:30μg), ceftazidime (CAZ:30μg), imipenem (IPM:10μg), meropenem (MEM: 10μg), piperacillin/tazobactam (PTZ: 100/10μg) and ampicillin/sulbactam (10/10μg) were placed on the medium. Then, the media were incubated for 18 hours at 37°C. The results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) recommendations.11 A control strain of Pseudomonas aeruginosa ATCC 27853 was used for quality control of susceptibility testing.
Phenotypic identification of ESBL producing isolates
Phenotypic identification of ESBL producing isolates have been carried out using disk potentiating test and confirmed by DDST screening method. Antibiogram disks containing CAZ (30μg), CTX (30μg), ceftazidime (30μg) + clavulanic acid (10μg) and cefotaxime (30μg) + clavulanic (10μg) were used. Pairs of disks (ceftazidime with ceftazidime/clavulanic acid and cefotaxime with cefotaxime/clavulanic) were placed on Muller-Hinton agar medium with 20 mm space between them. According to the CLSI criteria and manufacturer instruction, an increase in zone diameter of ≥5 mm in the presence of clavulanic acid indicated the presence of ESBL in the test organism. Klebsiella pneumoniae ATCC700603 was used as a positive control for ESBL production.11
Amplification of ESBL genes
The specific primers including CTX-M, SHV and TEM (Table 1) were used for PCR amplification of the genes. The PCR mixture contained the DNA template, the forward/reverse primers, and the master mix (Promega, USA). Amplification was carried out with the thermal cycling conditions according to the relevant studies and mentioned in Table 1. The PCR amplification was conducted in the thermal cycler device (Applied Biosystems, USA). Agarose gel electrophoresis of the amplified PCR products with 100bp size marker (Promega, USA) were carried out in a 2% agarose gel for 2h at 80V and stained with ethidium bromide. The PCR product bands were then checked under UV irradiation.
Primer name |
Primer sequence (5´ to 3´) |
Annealing temp. (°C) |
Product size (bp) |
Reference |
TEM |
F: ATGAGTATTCAACATTTCCG |
53 |
931 |
12 |
SHV |
F: AAGATCCACTATCGCCACAG |
60 |
231 |
13 |
CTX-M |
F: CGATGTGCAGTACCAGTAA |
60 |
585 |
14 |
Table 1 Primers used for PCR amplification of the ESBL genes
Antibiotic |
Number of sensitive (%) |
Number of intermediate (%) |
Number of resistant (%) |
Cefipime |
3 (7.5) |
0 (0.0) |
37(92.5) |
Cefotaxime |
1 (2.5) |
1 (2.5) |
38(95) |
Ceftazidime |
1 (2.5) |
2 (5) |
37(92.5) |
Imipenem |
6 (15) |
2 (5) |
32(80) |
Meropenem |
7 (6.6) |
4 (10) |
29(72.5) |
Piperacillin/tazobactam |
7 (17.5) |
3 (7.5) |
30(75) |
Ampicillin/sulbactam |
10 (25) |
4 (10) |
26(65) |
Piperacillin |
2 (5) |
2 (5) |
36(90) |
Table 2 The antibiotic resistance patterns of A. baumannii
In the current study 65 A. baumannii (72.2 %) were isolated out of 90 samples which collected from patients with burn infections in 2 hospitals of Baghdad City in Iraq. The isolates of A. baumannii were identified by using API 20NE system (bioMérieux) and the identification was confirmed by PCR amplification of the specific gene blaOXA-51 (Figure 1). A. baumannii isolates were identified by molecular detection of the carbapenemase gene blaOXA-51 in all 75 isolates, which then used to identify A. baumannii-mediated infections. Many previous studies indicated to the identification of A. baumannii at the species level by PCR detection of blaOXA-51-like genes [10, 15]. This study revealed that A. baumannii was one of the most important sources of nosocomial infections in ICUs and the prevalence of this species in ICU equipment in four hospitals in Baghdad city was 22.05%. ICUs are one of the units that have the highest occurrence rates of infections and the prevalence of infection is two to five times higher in ICUs than in the general inpatient population.16,17 Acinetobacter spp. are more frequently colonized on inanimate objects and hands of staff in the ICU than Pseudomonas spp and Staphylococcus aureus.18 The study conducted by Mehraban et al.18 revealed the high frequency of genus Acinetobacter among Gram negative bacteria isolated from environmental surfaces of ICUs of four hospitals located in the city of Qom, Iran.19 In another study in an adult ICU tertiary care Hospital in Riyadh, Saudi Arabia, A. baumannii was recognized as the most frequently isolated bacteria.20 Multidrug resistant A. baumannii is the most common cause of nosocomial infection in burn patients in Iran.21 Acinetobacter spp. can survive dry conditions for long periods, thus A. baumannii is frequently isolated from medical equipment and the contamination of hospital equipment caused by Acinetobacter outbreaks in the hospital environment is prevalent.22 Phenotypic detection of ESBL producing isolates of A. baumannii have been carried out using DDST screening method (Figure 2). From total of 65 isolates, 40 (61.53%) A. baumannii isolates were identified to be produced by ESBL enzymes. Susceptibility of 40 ESBL producing A. baumannii isolates to 8 of beta-lactam an antibiotic was evaluated by the Kirby-Bauer disk diffusion method (Table 2).The results showed high resistance to all antibiotics used in this study. Ninety-five percent, 92.5 %, 92.5 % and 90 % out of 40 A. baumannii isolates were resistant to Cefotaxime, Ceftazidime, Cefepime, and Piperacillin respectively. Also high resistance to Imipenem and moderate resistance to Meropenem, Piperacillin/tazobactam, and Ampicillin/sulbactam. Multidrug-resistant (MDR) A. baumannii became prevalent in many hospitals all over the world and has been recognized as one of the most important nosocomial pathogens especially in burn units.23 Our results showed high resistance of A. baumannii isolates to beta-lactam antibiotics such as penicillins, cephalosporins, and carbapenems used in the susceptibility test. Production of carbapenem-hydrolyzing beta-lactamases is one of the significant mechanisms of carbapenem resistance.24 The extended-spectrum beta-lactamases, ESBLs, play an important role in resistance against third-generation cephalosporins such as cefotaxime, ceftazidime, and cefepime.10 Specific primers were used to detect ESBL genes by PCR assay. Of all 40 A. baumannii isolates, 30 (75%), 18 (45%) and 11 (27.5%) isolates have been confirmed to harbor the blaTEM, blaCTX-M, and blaSHV genes, respectively. It was obvious that blaTEM genes were the most frequent ESBL types among A. baumannii strains in burn infections. PCR products of ESBL genes were analyzed by gel electrophoresis to detect the prevalence of these genes (Figures 3-5). In this study, the prevalence of ESBLs encoding genes among A. baumannii isolates has been investigated showing a high frequency of blaTEM genes among the isolates (75%). ESBL positive strains of A. baumannii and P. aeruginosa are increasingly found in hospital isolates. These strains revealed resistant to available antibiotics and these genes can transfer to other clinical strains. Molecular detection would determine which types of ESBL are present in each isolate and the identification of beta-lactamases would be essential for an epidemiological investigation of antimicrobial resistance.12 Ting et al.24 investigated the drug resistance genes in 7 strains of Imipenem- resistant A. baumannii including TEM, SHV, CTX-M, DHA, CIT, IMP, VIM, KPC, OXA-23, the results found TEM (100%) and OXA-23 (100%) genes among the isolates, but the other genes such as SHV, CTX-M, DHA, CIT, IMP, VIM, KPC could not be detected.25 The present findings was in contrast to the results of Khalilzadegan et al.25 who detected that the frequencies of blaTEM and blaCTX genes were 3.2% and 19.4%, respectively, while the level of resistance to various antibiotics was in the range of 69.4% to 100%.26 Another research reported that 40% of the isolates carried blaCTX and 12.8% carried blaTEM.26 The study of Odewale et al. in Ladoke Akintola University teaching hospital, Osogba, Nigeria, detected that 72.7% of patients in the ICU were most infected with A. baumannii and the isolates showed 90.9% resistance to both Ceftriaxone and Ceftazidime. The PCR results showed that blaTEM, and blaCTX-M genes were positive in some isolates, while blaSHV was not detected in any of the isolates. ESBL genes are plasmid-borne and are easily transmitted from one strain to another; this explains why these bacteria have firmly established themselves as MDR nosocomial pathogens whose infections no longer respond to treatment by commonly used antibiotics.14

Figure 1 Electrophoresis of the amplified products of blaOXA51 (353bp) genes by PCR in a 2% agarose gel. Lane 2-11: positive result of gene detection in A. baumannii isolates; Lane 12: negative result for A. lwoffii, Lane 1: negative control (PCR product without the DNA template); Lane M:100bp DNA ladder.
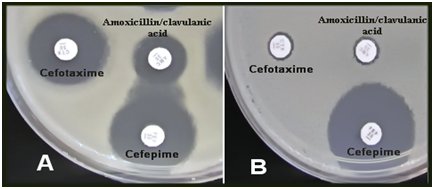
Figure 2 Double-disk synergy test for extended spectrum beta-lactamases in A. baumannii, A (positive result), B (negative result). AMC – Amoxycillin + Clavulanate, CTX-Cefotaxime, FEP–Cefepime (A clear extension of the edge (Synergy) of the FEP inhibition zone towards the disc containing clavulanate).
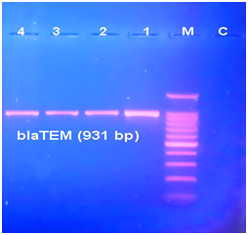
Figure 3Electrophoresis of the amplified products of blaTEM (931bp) genes by a PCR in a 2 % agarose gel. Lane 1-4: positive result of gene detection in A. baumannii isolates; Lane C: negative control (PCR product without the DNA template); Lane M: 100bp DNA ladder.
The presence of A. baumannii in ICU increases the risk of transmission to patients leading to nosocomial infections. The prevention of the nosocomial infections needs the quick detection of the sources of contamination, especially on the equipment. Detection of ESBL genes by PCR is known as an effective and sensitive method for epidemiological analysis and tracing MDR A.baumannii. ESBL producing bacteria should be identified quickly so that appropriate antibiotic usage can be conducted.
My Institute’s (Institute of Genetic Engineering and Biotechnology for Postgraduate Studies) representative is fully aware of this submission.
The author declares there is no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.