MOJ
eISSN: 2374-6912


Targeted gene modifications to animals’ especially mammalian genomes are central to understand the role of the gene in development, physiology, and different disease conditions. Functional elucidations of a gene or genetic variants require precise editing technologies. In conventional gene targeting methods, mutations of the gene or gene has been introduced into the genome by homologous recombination in embryonic stem (ES) cells and then injected into wild-type Blastocyst to generate chimerical animal models containing the targeted gene modification. Although it is widely used approach, however, it is laborious, time-consuming, and expensive, needing well-trained researchers. Moreover, in most of the mammalian species, established ES cell lines are not available to produce chimeric animals, which limits the traditional approach. The recent discovery of new genome-engineering tools, especially CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/ CRISPR-associated) overcome much of the difficulties. CRISPR is a microbial nuclease system that is involved in defense against genome invading phages and plasmids. It has been experimentally proven that the Cas9 end nuclease from Streptococcus pyrogens type II CRISPR/Cas system can be programmed to produce sequence-specific double strand breaks (DSBs) in vitro by providing a synthetic guide RNA (gRNA-fusion of crRNA and tracrRNA). There are lots of researchers that have illustrated that the CRISPR/Cas9 system can be used to generate different gene deletions or replacement in mammalian as well as in model organisms. In this review, we will discuss the pros and cons of the system and briefly discuss the future direction.
Keywords: crispr/cas9 technology, molecular scissors, recombinant dna technology, insertional mutagenesis, genes mutations
ES, embryonic stem; DSBs, double strand breaks; NHEJ, non-homologous end joining; gRNAs, guide rnas; TALEN, transcription activator-like effector nuclease; ZFN, zinc finger nucleases; tracrRNA, trans-activating crrna; crRNAs, crispr rnas
The understanding of the mammalian genome has increased significantly as recombinant DNA technology has grown ever faster and cheaper over the last decade. Modulation of the genome both in vitro and in vivo is much easier now. In vivo gene modification to create transgenic animals are valuable tools for studying development and disease. Transgenic animals represent a crucial model to study cell biology, genomics, proteomics, and many other applications in biotechnology and basic science. Transgenic animals are conventionally generated by insertional mutagenesis1,2 or by gene-targeting methods.3 However, these methods rely on the random integration of the transgene into the genome followed by drug selection. To identify cell populations expressing the transgene is a laborious screening process in the traditional method. In the recent decade, there have been a few methods developed which expedite the pace of genome editing technology. They are ZFN (Zinc Finger Nucleases), TALEN (Transcription Activator-Like Effector Nuclease) and the type II bacterial CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/ CRISPR–Associated) technologies.4–8 Among them, CRISPR/Cas9 technology is considered as the best efficient gene-editing technology as the efficiencies of ZFN and TALEN are low.9,10 Also, ZFNs and TALENs remain somewhat difficult and expensive to design, develop, and empirically test in the cellular context. Further, with CRISPR/Cas9 technology, it is easy to design and delivery of multiple gRNAs suggests the possibility of multiplexed genome editing in a short time.
CRISPR/Cas9 is an RNA-guided immune system that protect Archaea and Bacteria cells against genome invading viruses and plasmids.11,12 The puzzling CRISPR DNA segments were first observed in 1980, but their function was not well understood for almost two decades.11 In general, when an unknown virus attacks, bacteria incorporate viral DNA sequences into the repetitive sequences of their genome. Thus, when the bacteria again encounter that virus, bacteria use the DNA in these repetitive sequences to synthesize RNAs that recognize the matching viral sequences. Next, a protein attached to one of these RNAs cut up the viral DNA. Two forms of gRNAs are used to facilitate Cas9-mediated DNA cleavage, these are a Crispr RNA (crRNAs) array and trans-activating crRNA (tracrRNA) (Figure 1). This crRNA in association with tracrRNA guide the Cas9 protein to a region called the ‘protospacer adjacent motif’ (PAM), a three-nucleotide sequence (NGG) juxtaposed to the DNA complementary region in the DNA (Figure 1).5,13,14 In the presence of an appropriate homologous donor, the CRISPR/Cas9 system may be used to generate defined modifications and insertions at a targeted locus through the homologous DNA repair process. In the absence of a donor, single DSBs generated by CRISPR/Cas9 are repaired through the error-prone non-homologous end joining (NHEJ), which is prone to insertion or deletion mutations.
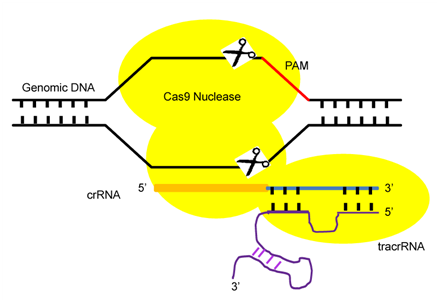
Figure 1 The components of CRISPR/Cas9 System.
Pictorial Diagram showing the three components:
Recently, several research groups have been adapted CRISPR/Cas9 system for site-specific genome editing in diverse cell types and organisms such as zebrafish,15–17 Drosophila,18 mice19–21 and human cells.9,22–24 A recent report also demonstrated the disruption of a Green Fluorescent Protein transgene in mice using the CRISPR/Cas9 system.19 Yang et al.20 created reporter and conditional mutant mice by co-injection of zygotes with Cas9 mRNA and different guide RNAs (gRNAs) as well as DNA vectors of different sizes. This group generated mice carrying a fluorescent reporter construct with different genes.20 They also analyzed potential off-targets of five gRNAs in gene-modified mice and ESc lines and identified off-target mutations are rare.20 In a recent study, Zheng et al.21 used two gRNAs coupled with Cas9 and efficiently created DNA deletions of 10kb. Targeted deletion with CRISPR/Cas9 observed to be independent of the transcriptional status of the targeted locus.21
Tools for genome editing seem to be improving faster than software technologies. Recently developed genomic editing technologies have the potential to generate different transgenic animals because of their ability to modulate genes. The pros and cons of different genome editing technologies are summarized in Table 1. CRISPR/Cas9 systems are found to be comparable to ZFN and TALEN regarding activity but cheaper and ease to use.9,24–26 Mashiko et al.26 generated mutant mice by injecting humanized Cas9 (hCas) mRNA and single guide RNA into fertilized eggs. The same group also produced mutant mice by injecting circular plasmids expressing Cas9 instead of Cas9 mRNA and sgRNA into mouse zygotes. Interestingly, multiple gRNAs can be used to edit multiple sites within the genome, demonstrating the one-step generation of animals carrying multiple genes mutations. Zheng et al.21 showed that CRISPR/Cas-mediated gene editing could disrupt multiple genes in mouse ES cells with 80% efficiency. Thus, CRISPR/Cas technology greatly accelerate the in vivo study of functionally redundant genes and of epistatic gene interactions.
Present Gene Editing options |
Details |
Advantage |
Disadvantage |
Zinc Finger Nuclease (ZFN) |
A protein consisting of a DNA-cutting enzyme and a DNA-grabbing region that can be tailored to recognize different Genes |
It was the first programmable genome-editing tool and faster and cheaper than the traditional methods. |
|
TALEN (Transcription Activator-Like Effector Nuclease) |
Protein is containing a DNA-cutting enzyme and a DNA-grabbing region that can be programmed to recognize different genes. |
This is easier to design than ZFN. It also is cheaper than ZFN. |
|
CRISPR/Cas9 |
A DNA-cutting protein guided by an RNA molecule that can find the specific gene of interest. |
This technique is affordable, easy to use, and it works for high throughput multi-gene experiments. CRISPR also give scientists a precise way to delete and edit specific bits of DNA-even by changing a single base pair. |
It can make off-target cuts. However, improved engineered Cas9 protein reduces the chance off-target cuts. |
Table 1 Clinical and biochemical variables of individuals with overweight-obesity
SD: Standard Deviation; BMI: Body Mass Index; WC: Waist Circumference; AC: Abdominal Circumference; HC: Hip Circumference; RER: Respiratory Exchange Ratio; HR: Hear Rate.
The genome editing technology is advancing at an exceptionally rapid pace but there remains some key issues regarding development of appropriate assays to evaluate off-target effects and establish safety. Thus, off-target errors are the greatest disadvantage of CRISPR/Cas9 system. Recently, different research groups optimized CRISPR/Cas9 component to drive down its error rate.27–30 Different genetically altered Cas9 enzyme were synthesized that reduced off-target errors at least tenfold compared with unaltered Cas9 enzymes.31 In this direction, various groups are working to reduce further off-targets cut by Cas9 enzyme.
Animal models are widely used to understand the complex process of development and disease. Reporter transgenic animals are critical tools to study specific cells or organs. As CRISPR-Cas9 is so quick, cheap and easy to use, the creation of more and more reporter transgenic animals will help us to understand the development and disease more precisely. One of the most important things of this technology is to make multiple genetic changes at once. In this context, the study of multiple genes at a single time in the same animal is possible. Further, this technology might be utilized for the complex human genetic disease such as autism, sickle-cell anemia, HIV, cystic fibrosis or schizophrenia. However, there remains a significant amount of research that needs to be done in order to utilize CRISPR/Cas9 for the benefits of the patients. We still do not to know what the potential effects and consequences could be if we correct the mutated DNA in sickle-cell patient’s stem cells and put back into the patient; what will be the consequences. However, improvement of this technique could be deployed in the coming future to treat deadly genetic diseases. In this direction research works are going on and hopefully, we will get an answer soon.
In summary, CRISPR/Cas9 technology is an efficient and simple method of generating sophisticated genetic modifications in one step. We need to take maximum caution for therapeutic application of this technology to make sure that the enzyme does not cause unwanted genetic modifications. Recently, researchers from the United States of America, United Kingdom, and China were convening in Washington DC to discuss the different ethical issues of gene editing technologies.30
I thank Dr. Keigo Machida (Associate Professor, University of Southern California, CA) for introducing me this technology for my ongoing transgenic mouse work.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.