MOJ
eISSN: 2374-6912


In our days it becomes clear that not only the genetic information encoded in the sequence of DNA is responsible for gene expression profile but the epigenetic signature encoded in the pattern of modifications of histones that associate with DNA in chromosomes also plays a very important role in the determination of gene expression. Octomere of histones (H3/H4 and H2A/H2B) and a segment of DNA surrounding histone proteins compose a nucleosome, a basic unit of chromatin that in turn is the first level of chromosome organization. Besides the hypermethylation of CpG islands of DNA that leads to gene repression multiple modifications of histones also influence gene expression either activating or inhibiting it. These histone modifications altogether are called a “histone code” and include such diverse covalent modifications of Lys, Arg, Ser residues of histones as acetylation, methylation, phosphorylation, ADP-ribosylation, ubiquitination that are usually reversible. While acetylation of histones is an activating mark, methylation of histones at particular residues could be either repressive or activating modification. Although changes in histone modification pattern already lead to changes in gene expression, the data were accumulated indicating that complete replacement of whole nucleosome or nucleosome arrays with newly synthesized and modified nucleosomes could induce gene activation or repression. In this review the data on different epigenetic mechanisms of gene expression regulation will be presented. Regulation of interferon-beta gene expression will be described in more details as comprehensively studied example of epigenetic regulation of gene expression.
Keywords: gene expression, epigenetic regulation, nucleosome, histone code, interferon-beta gene
HAT, histone acetyl transferase; HDAC, histone deacetylase; HMT, histone methyltransferase; HDM, histone demethylase; IFN-Beta, interferon-beta; RC, replication-coupled; RI, replication-independent; TSA, trichostatin a
DNA replication involves the chromatin assembly on new DNA strands which involves the formation of new nucleosomes, the basic unit of chromatin. Nucleosome is composed of the tetramer of H3/H4 histones and two dimers of histones H2A/H2B that are wrapped around with DNA segment.1,2 H3/H4 and H2A/H2B histones are called core histones as they are positioned in the center of nucleosome. Some core histones are presented by one isoform such as histone H4, while other histones exist in several isoforms such as histone H3 (H3.1, H3.3). Histone H3 is of special interest as its different isoforms are involved in diverse key nuclear processes: H3.1 isoform is involved in DNA replication and repair maintaining the genome integrity and H3.3 isoform is implicated mainly in transcription which comprises the first step of gene expression.1,2
In our days it became clear that not only the sequence of DNA by itself determines gene expression profile but also the epigenetic modifications of DNA sequence that include the methylation of CpG islands3 and epigenetic modification of histones composing the nucleosomes play a very important role in gene expression regulation. In this review I will discuss mainly the epigenetic regulation of gene expression at the level of histone modifications and nucleosome assembly. As an example of such epigenetic regulation I will present the data on comprehensively studied regulation of interferon-beta (IFN-beta) gene expression.
Combinatorial nature of histone modifications
Gene expression in the context of chromatin is very complex process depending on DNA sequence and epigenetic modifications of DNA and chromatin proteins, mainly histones.1–3 Most common reversible modifications of histones H3 and H4 include acetylation of Lys (K) residues, methylation of Lys (K) and Arg (R) residues and phosphorylation of Ser (S) residues mainly on N-terminus of histone molecules and they are presented schematically at (Figure 1). These combinatorial sets of histone amino-terminal modifications were given the name “histone code” in the beginning of our century4 introducing additional informational system of modified proteins bound to DNA and, therefore, remarkably extending the information potential of the genetic code discovered long before. Since the introduction of the definition “histone code” in 2001 the literature in this field (research and review articles) is growing in exponential progression.

Figure 1 Most common histone reversible modifications and modifying enzymes.
Reversible covalent modifications, particularly characteristic for histones H3 and H4 that compose the nucleosome, the basic unit of chromatin, and corresponding enzymes are presented. These covalent modifications include acetylation of Lys (K) residues, methylation of Lys (K) and Arg (R) residues and phosphorylation of Ser (S) residues of histones. For more detailed explanation see text.
Nucleosome replacement as one of gene activation mechanisms
In addition to covalent modifications of individual histone molecules accumulating data indicate that gene activation could be accompanied with whole nucleosome replacement at promoters and coding regions.5–8 Several works indicate that gene activation is accompanied by the incorporation of histone replacement variant H3.3.7,8 Moreover Schwartz and Ahmad5 have shown that this acquirement of H3.3 is accompanied by the loss of canonical histone H3 in Drosophila (which corresponds to H3.1 in mammals). Besides this, heterochromatin protein 1 (HP1), which binds to H3meK9 and is involved in heterochromatin formation and gene silencing, shows dynamic behavior in mammalian cells suggesting the possibility of nucleosome replacement.9,10 Nucleosome replacement could be necessary to remove irreversibly modified histones or slowly demethylated histones. While acetylation and phosphorylation marks could be removed by histone deacetylases and dephosphotases, histone methylation for a long time was considered irreversible modification. This view on the irreversibility of histone methylation was changed since first decade of our century when several histone demethylases (HDMs) has been identified belonging to the Jumonji (Jmj) family of demethylases that epigenetically regulate gene expression through the removal of methyl groups from histones and have varying specificities for lysine site and the degree of methylation of the substrate and for chromatin context.11–13 Although the number of newly discovered HDMs grows further it seems that methylation of histone H3 is more stable than other histone modifications such as acetylation and phosphorylation and one of the remaining ways to remove it is to replace the methylated histone molecules with non-methylated ones implying also the replacement of the whole nucleosome. The enrichment of the replacement histone H3.3 variant with modifications associated with active chromatin14 also suggests that upon activation of transcription histone H3.1 is replaced with H3.3.
Epigenetic mechanisms of the regulation of IFN-beta gene expression
Epigenetic mechanisms of the IFN-beta gene activation
Activation of interferon-beta (IFN-beta) gene takes place during multiple cellular processes including antiviral and immune responses.15,16 IFN-beta gene expression after viral infection is a comprehensively studied example of gene induction involving multiple epigenetic modifications of histone proteins.17,18 Firstly, the enhanceosome is assembled at promoter region of IFN-beta gene. Further, the enhanceosome recruits chromatin modifying (histone-acetyltransferases, HATs) and remodeling enzymes of SWI/SNF family, PolII as well as general transcription factors what finally leads to the activation of transcription. In many cases of gene activation the acetylation of histones and nucleosome remodeling are the main causes of gene induction. Agalioti and co-authors17,18 showed that upon activation of IFN-beta gene HAT GCN5 modifies several lysines that mediates the recruitment of the SWI/SNF complex and TFIID and finally activates transcription. Using ChIP technique they showed that several lysines (H4 K8, H3 K9) start to be acetylated 3 hrs after viral infection. Although the activation of IFN-beta gene expression described above is studied in details still some questions remain unanswered concerning the mechanisms of the transition from repressed to activated state and the maintenance of repressed and activated states.
Mechanisms of the establishment and maintenance of the IFN-beta gene repressed state
Several mechanisms could be involved in the establishment of repressed state of IFN-beta gene. Firstly, histone deacetylation is involved in the establishment of constitutive repressed state,19 which is reversed by histone acetylation upon the activation of IFN-beta promoter. One of the possible candidates which could recruit a histone deacetylase (HDAC) activity to the IFN-beta promoter is the transcription factor YY1. We have shown that this transcription factor, capable to interact with both HATs and HDACs, could regulate the IFN-beta gene expression via its dual activator/repressor activity.20 It has been demonstrated also that PRDI-BF1, a protein involved in post-induction repression of IFN-beta gene transcription, recruits the histone methyltransferase (HMT) G9a to IFN-beta promoter to induce its silencing via methylation of H3 K9.21 As it was indicated earlier the demethylation of H3K9 could be slower or less efficient than removal of other histone modifications. Although multiple HDM were discovered11,12 and could be involved in the demethylation of H3meK9 at the IFN-beta promoter upon its activation, still the possibility exists that upon activation of this promoter the methylated histones could be replaced by acetylated ones via nucleosome replacement.
It is not clear when the methylation mark could be removed from the IFN-beta promoter, long before its activation or upon its activation during the assembly of the enhanceosome, and whether the nucleosome replacement is involved in the methylation mark removal. The fact that the first acetylations at the IFN-beta promoter take place starting 3 hrs after induction17 which correlates with the time found for the incorporation of H3.3 (bearing the modifications associated with active chromatin) in gene array upon its activation7 also argues in favor of the nucleosome replacement at IFN-beta promoter. The work of Schwartz & Ahmad5 demonstrates that H3.3 deposition spreads over large transcription units indicating that nucleosome replacement takes place during transcription elongation as well. The possibility could not be ruled out that the H3meK9 spreads along IFN-beta gene beyond the promoter region upon post-induction repression. In this case the nucleosome replacement would be necessary at the promoter region upon activation of transcription and along the whole IFN-beta transcription unit during transcription elongation.
Histone predeposition complexes as prerequisites of nucleosome assembly and replacement Predeposition complexes containing histone variants H3.1 and H3.3
In cellular environment histone predeposition complexes are involved in the formation of mature nucleosome incorporated in the chromatin structure. These histone predeposition complexes containing histones and associated proteins start to form in the cytoplasm then they are transported into the nucleus through the nuclear pores and incorporate there into the nucleosome. Histone predeposition complexes containing histone variants H3.1 and H3.3 were purified and extensively characterized. These studies shed light on the mechanisms of nucleosome assembly and nucleosome replacement in chromatin. The complex containing the histone chaperone CAF-1 and histone variant H3.1 is involved in the replication-coupled (RC) histone deposition during S-phase while the complex containing histone chaperone HIRA and histone variant H3.3 participate in the replication-independent (RI) histone deposition which occurs during all stages of the cell cycle.22–24 All these complexes contain dimers and not tetramers of H3/H4. Histone chaperone ASF1A/B is a component of both complexes. Crystallographic studies of ASF1 bound to H3/H4 dimer25,26 or of N-terminal domain of ASF1 bound to C-terminal helix of histone H327 showed that ASF1 binds to histone H3 in an orientation that prevents the interaction of two histones H3 and therefore H3/H4 tetramer formation. These investigations explain the discrepancy between the fact of spontaneous H3/H4 tetramer formation from pure histones H3 and H4 in vitro28,29 and the existence of H3/H4 dimers in histone predeposition complexes containing ASF1 in nucleoplasm and cytoplasm.22–24,30
Characterization of cytoplasmic and nuclear histone predeposition complexes
In a recent study a comprehensive characterization and comparison of cytoplasmic and nuclear histone H3.1 predeposition complexes was carried out.24 To reveal the precise compositions of cytoplasmic and nuclear H3.1 histone predeposition complexes we purified them from transduced HeLa cells expressing Flag/HA-tagged histone H3.1 using the method of tandem affinity purification, determined the protein composition with mass spectrometry approach and confirmed the results of mass spectrometry analysis with Western blotting technique. The comparison of cytoplasmic and nuclear H3.1 histone predeposition complexes demonstrated similar composition of histone H3.1 predeposition complexes present in two main compartments of the cell, cytoplasm and nucleus, although several prominent differences were observed. The major differences that we revealed in these complexes were the following: we identified the importin-beta 3 in cytoplasmic complex which was not found in nuclear one and we demonstrated the presence of the large subunit of histone chaperone CAF-1 p150 only in the nuclear complex (Figure 2, this figure with no.1 will be published in the article accepted by the Journal of Investigative Genomics in volume 2, issue 1: Shestakova EA., Nakatani Y. (2015) Characterization of histone predeposition complexes from different cellular compartments, see also the Ref. no. 24).

Figure 2 Mass spectrometric and immunoblotting analyses of nuclear and cytoplasmic H3.1 histone predeposition complexes.
(A) Silver staining of the H3.1 nuclear (lane 1) and cytoplasmic (lane 2) predeposition complexes obtained by tandem affinity purification on anti-Flag and anti-HA antibodies coupled agarose beads from the extracts of transduced HeLa cells. The proteins identified by mass spectrometric analysis are indicated. (B) Immunoblot analysis of the H3.1 nuclear (lane 1) and cytoplasmic (lane 2) predeposition complexes. Complexes were analyzed by immunoblotting with anti-CAF1p150, anti-ASF1A/B, anti-CAF1p48 (RbAp48), anti-Mcm2, anti-HAT1, anti-importin beta 3, anti-HA antibodies. The latter antibody was used to reveal the epitope-tagged histone H3.1.
Histone acetylation/deacetylation as a possible mechanism of modulation of H3/H4 tetramer versus dimer formation
Experiments with TSA, a histone deacetylase inhibitor, demonstrated that increased acetylation of histone H3.1 and possibly other proteins allowed the formation of H3/H4 tetramer instead of H3/H4 dimer in cytoplasm and nucleoplasm.24 In relation with this it is worth to note the article by Adkins et al.31 demonstrating that the binding of Asf1 to histones induces the acetylation of several residues of newly synthesized histone H3 (lysine 9 and lysine 56) by GCN5 and other HATs in S-phase of cell cycle probably alleviating further histone assembly in tetramer onto newly replicated DNA.
Model of successive formation of cytoplasmic and nuclear histone predeposition complexes leading to nucleosome assembly in chromatin
These findings allowed us to suggest the model according to which the core complex is formed in the cytoplasm that contains the dimer of histones H3/H4, the histone chaperone ASF1A/B, the histone acetyltransferase HAT1, the small subunit of histone chaperone CAF-1 p48, MCM complex involved in the replication of DNA and importin-beta 3. With the help of the importin-beta 3 the core complex is transported into the nucleus through the nuclear pore, loses the importin-beta 3 on the inner side of the nuclear pore and acquires additional subunits of histone chaperone CAF-1 (p150 and p60) forming the complete histone H3.1 predeposition complex that assembles the nucleosome on the replicating DNA (Figure 3, this figure with no.5 will be published in the article accepted by the Journal of Investigative Genomics in volume 2, issue 1: Shestakova EA., Nakatani Y (2015) Characterization of histone predeposition complexes from different cellular compartments, see also the Ref. no. 24). Similar mechanisms could be suggested for the formation of predeposition complex containing histone H3.3 and its incorporation into chromatin potentially leading to nucleosome replacement upon gene activation.
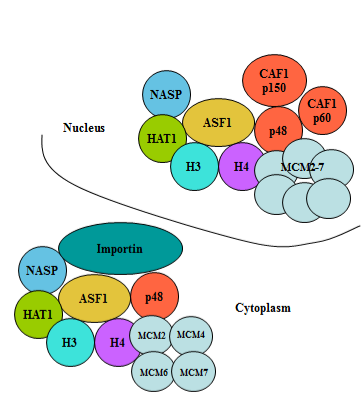
Figure 3 Model of histone H3.1 predeposition complex formation from cytoplasm to nucleus.
According to this model the core complex is formed in the cytoplasm that contains the dimer of histones H3/H4, the histone chaperone ASF1A/B, the histone acetyltransferase HAT1, the small subunit of histone chaperone CAF-1 p48, MCM complex involved in the replication of DNA and importin-beta 3. With the help of the importin-beta 3 the core complex is transported into the nucleus through the nuclear pores, loses the importin-beta 3 on the inner side of the nuclear pore and acquires the additional subunits of histone chaperone CAF-1 (p150 and p60) forming the complete histone H3.1 predeposition complex that assembles the nucleosome on the replicating DNA.
Taken together, these series of studies not only explains the mechanisms of the assembly and the replacement of basic unit of chromatin, the nucleosome, but also opens up vast possibilities to develop new medicaments directed against epigenetic targets in oncological diseases. This direction of research is developing rapidly in our days. One of the best examples of such investigations is the development of very specific inhibitors ((+)-JQ1) of BET bromodomain-containing chromatin adaptor protein subfamily involved in the recognition of acetylated histones.32 These inhibitors abrogate the MYC gene transcription resulting in significant anti-tumor activity in xenograft models of Burkitt’s lymphoma and acute myeloid leukemia.33 Therefore, these inhibitors have perspective clinical utility as MYC oncogene is a very powerful inducer of different malignancies including Burkitt’s lymphoma and acute myeloid leukemia. Moreover, the combination of HDAC inhibitor, vorinostat, and hormone therapy is becoming a very powerful approach in breast cancer epigenetic therapy.34 HDAC inhibitors as well as DNA methyltransferases inhibitors exist already for considerable time and belong to first generation epigenetic inhibitors. Generally speaking different histone post-translational modifications together with DNA methylation constitute an “epigenetic code”35 and aberrant changes of this code are very perspective and promising targets of anti-cancer therapy in our days.
For example, apart acetylated histone marks, many other chromatin epigenetic modifications such as methylated residues of histones and enzymes involved in these histone modifications arise as very important targets for drug discovery.12 In particular, many HMT and HDM have become promising candidate oncology targets in recent years.13,35 HMT inhibitors including inhibitors of G9a, EZH2, DOT1L and HDM inhibitors including LSD1 and Jumonji C inhibitors represent second generation epigenetic inhibitors.35 Therefore, these studies point the new directions and roads in the development of medicaments targeting the epigenetic mechanisms of cancer initiation and progression therefore improving targeted anti-cancer therapy. Intensive and extensive introduction of such medicaments in clinics would be one of the promising ways in the eradication of cancer, one of the most harmful diseases of our days.
The author (Dr. Elena A. Shestakova) is grateful to Dr. Yoshihiro Nakatani (Dana-Farber Cancer Institute and Harvard Medical School, Boston, USA) in whose laboratory the studies cited in this review were carried out. This research on histone predeposition complexes was supported with grants from NIH (GM065939-02) and Human Frontier Science Program to Dr. Yoshihiro Nakatani. Dr. Elena A. Shestakova holds a position of Senior Research Scientist, PhD at Blokhin Russian Cancer Research Center, Moscow, Russian Federation.
The author declares no conflict of interest.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.