MOJ
eISSN: 2374-6912


Nucleocytoplasmic shuttling of signaling molecules is crucial for regulating their activity. Regulation of signal transducer and activator of transcription 3 (STAT3) is critical for normal physiology while STAT3 dysregulation underlies many diseases such as cancer. However, the mechanism(s) underlying STAT3 nuclear egress remains unclear. Here, we show that exportin 7 is critical for STAT3 nuclear egress. Lysine acetylation at K685, frequently found in tumors and tumor cell lines, mediates STAT3 engagement with exportin 7. Blocking acetylation through drug administration or mutation of lysine K685 disrupted STAT3’s physical interaction with exportin 7, leading to STAT3 nuclear retention. Inhibition of STAT3 lysine acetylation significantly altered the functional localization of exportin 7 from the cell cytoplasm toward the nucleus which can be reversed by treating tumor with a lysine-acetylated peptide spanning STAT3 K685. Taken together, our results have identified exportin 7 as an essential karyopherin for STAT3 nucleocytoplasmic shuttling.
Keywords: stat3, exportin 7, nucleocytoplasmic shuttling
STAT, signal transducer and activator of transcription; NPC, nuclear pore complex; NES, nuclear export sequences; FPP, fluorescence protease protection
For a signaling molecule to function effectively it is necessary to have precise cellular localization. Consequently, it is critical to understand the regulation of cellular trafficking- both nuclear translocation and egress of signaling molecules. Signal transducer and activator of transcription 3 (STAT3) plays an important role in normal physiology and in several diseases especially cancer.1–4 Activation of STAT3 has been suggested to facilitate its nuclear accumulation.5–7 Nuclear STAT3 binds to DNA and regulates gene expression. STAT3 then dissociates from DNA and exits the nuclear compartment through the nuclear pore complex (NPC) into the cytoplasm, where it becomes a substrate for reactivation by kinases residing in the cell cytoplasm or at the outer cell membrane.8–10 The reactivation is necessary for STAT3 nuclear retranslocation and transcriptional activity.
Nucleocytoplasmic shuttling of transcription factors is tightly regulated by karyopherins. Karyopherins are a super-family of factors comprising importins and exportins responsible for facilitating trafficking of cargoes through the nuclear pore complex, thereby maintaining cargoes’ compartmental balance.11 While STAT3 nuclear translocation has been shown to be mediated by importin-α3 and importin-β1,12,13 the process that facilitates STAT3 nuclear egress remains poorly understood.7 Exportins (exportin 1-7), are thought to engage specific cargoes given by recognition of consensus motifs (nuclear export sequences, NES).14 Once exportins have shuttled their cargoes through the NPC in a RanGTP- dependent fashion, cargo molecules are thought to be released upon GTP hydrolysation in the cytoplasm.15,16 Important previous studies identified exportin 1/Crm1 to be critical for nuclear export of STAT1,17–19 emphasizing a NES that has a potential homologue in STAT3. Nevertheless, a more detailed analysis based on site-directed mutagenesis in STAT3 suggested that STAT3 nucleocytoplasmic shuttling may not involve Crm1.7,20 It remains unclear what exportin(s) is critical for STAT3 nuclear egress. In the current study, we investigated STAT3 nuclear egress in cultured cells as well as in tumors in vivo.
Mice and cell culture
For subcutaneous tumor challenge, athymic nude/nude mice (NCI Frederick) were injected s.c. into the flank with 5x106 MEFStat3D/D cells stably reconstituted with either STAT3wt-YFP or STAT3K685R-YFP. Subcutaneously engrafted MEFStat3D/D expressing STAT3-K685R-YFP were treated after tumors reached 5-7mm in diameter with 10mg peptide/mouse delivered by biolipid-carrier (BioPORTER Protein Delivery Reagent, GenLantis, San Diego, CA) according to manufacturer’s instructions. Mouse care and experimental procedures with mice were performed under pathogen-free conditions in accordance with established institutional guidance and approved protocols from the Research Animal Care Committees of the City of Hope.
Stat3-deficient mouse embryonal fibroblasts (MEFStat3D/D, kind gift of Dr. Valeria Poli, Molecular Biotechnology Center, University of Torino, Torino, Italy), mouse 3T3/v-Src fibroblasts (gift of Dr. Richard Jove, The Vaccine & Gene Therapy Institute of Florida, Port St. Lucie, FL) and human glioma U251 cells (provided by Dr. Christine Brown, City of Hope, CA) were cultured in DMEM medium (Gibco) supplemented with 10% FBS (Sigma). Human prostate adenocarcinoma cells PC3, human prostate carcinoma DU145 and human breast adenocarcinoma MDA-MB468 were purchased from ATCC (American Type Culture Collection, Manassas, VA) and grown according to vendor’s instructions. Human colon carcinoma HCT cells with STAT3-wt and mutated STAT3-K685R (kind gift of Dr. Zhenghe Wang, Case Western Reserve University, Cleveland) were grown in McCoy’s 5A media supplemented with 10% FBS (Sigma).
Imaging
Indirect immunoflourescence was carried out as described previously,6 staining pY705-STAT3, STAT3, exportin 7, nucleoporin 50 and 153 (Santa Cruz), Hoechst33342 (Sigma), acetylated STAT3-K685 (Cell Signaling Technology), CD31 (BD Biosciences), and Ki67 (abcam). Fluorescence protease protection (FPP) assay and iFLAP analyses were carried out as described previously.8,21 Nuclear accumulation of proteins was calculated as described previously.22 The DuoLink® assay to localize interacting proteins was performed according to manufacturer’s instructions (Olink Bioscience, Goteborg, Sweden) using antibodies against Stat3 (Stat3 C-20, Santa Cruz) and exportin 7 (Acris). For control, a Stat3 blocking peptide (C-20 Stat3 blocking peptide, Santa Cruz) was added. Tissue array was purchased from BioMax. All specimens were analyzed by confocal microscopy (Zeiss LSM510).
Immunoblotting and immunoprecipitation
Whole cell lysates were prepared using RIPA lysis buffer containing 50mM Tris (pH 7.4), 150mM NaCl, 1mM EDTA, 0.5% NP-40, 1mM NaF, 15% glycerol, and 20mM β-glycerophosphate. Protease inhibitor cocktail was added fresh to lysis buffer (Mini Protease Inhibitor Cocktail, Roche). Normalized protein amounts were subjected to Western blotting, and immunodetection was performed using antibodies against STAT3, exportin 1, 2, 5, 7 (Santa Cruz), exportin 3 (Millipore), exportin 4 (Epitomics), exportin 6 (Proteintech), pY705-STAT3, acetylated STAT3-K685 (Cell Signaling Technology). Resveratrol was obtained from Cayman Chemical. For co-immunoprecipitation, anti-STAT3, exportin 7, nucleoporin 50 and 153 antibodies (Santa Cruz) were used to label rProtein G agarose beads (Invitrogen), incubated over night with lysates and Western blot detection was performed.
Polymerase chain reaction
Transcript amplification was determined from RNA purified using RNeasy Kit (QIAGEN). cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad). Real-time PCR was performed using Chromo4 Real-Time Detector (Bio-Rad). Murine and human Gapdh housekeeping genes were used to normalize target gene mRNA levels. Commercially available primers for analyses of STAT3 target genes and XPO7 were obtained from SA Biosciences/QIAGEN (Valencia, CA).
Exportin 7 mediates STAT3 nuclear egress via interaction with lysine acetylation
To identify the karyopherin(s) that regulates STAT3 nuclear export, we carried out STAT3 immunoprecipitation followed by western blotting with indicated exportin antibodies, using protein extracts prepared from human glioma U251 cells. Exportin 7, but not exportins 1-6, was in complex with STAT3, suggesting that exportin 7 might be involved in STAT3 nuclear egress (Figure 1A). Moreover, we determined STAT3 and exportin 7 interaction mainly localized in the cell cytoplasm (Figure 1B). STAT3 interaction with exportin 7 was further shown in tumors in vivo by co-immunoprecipitation using homogenates from human glioma U251 tumors from xenograft mouse models (Figure 1C).
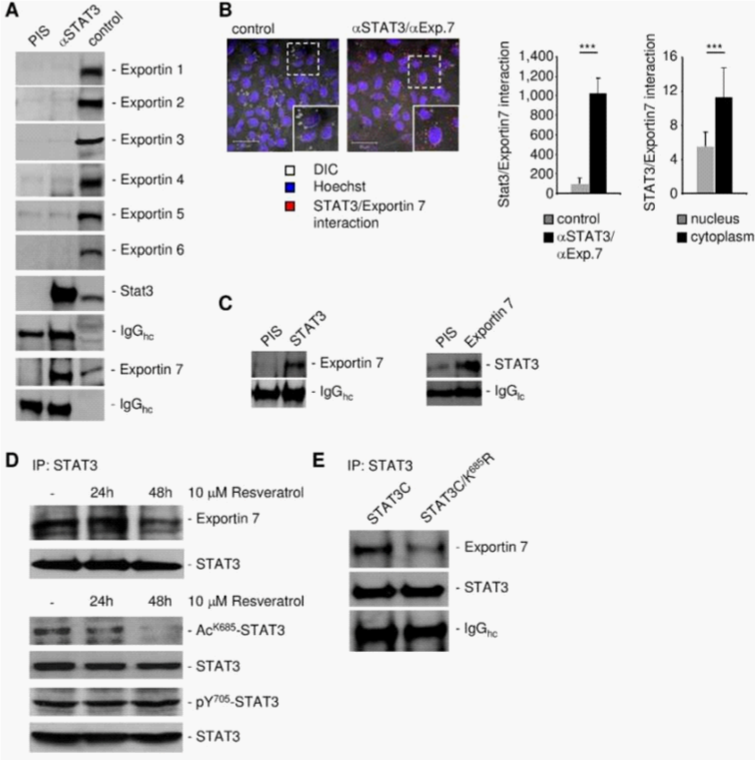
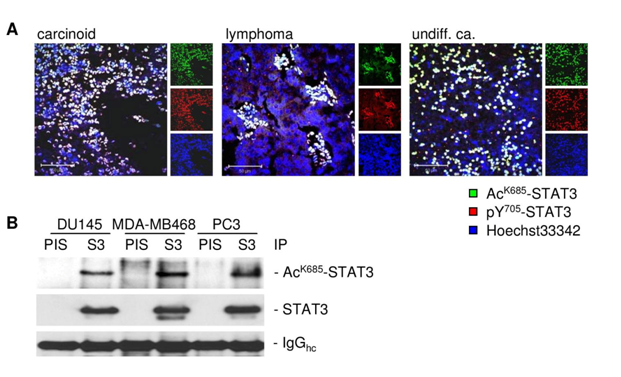
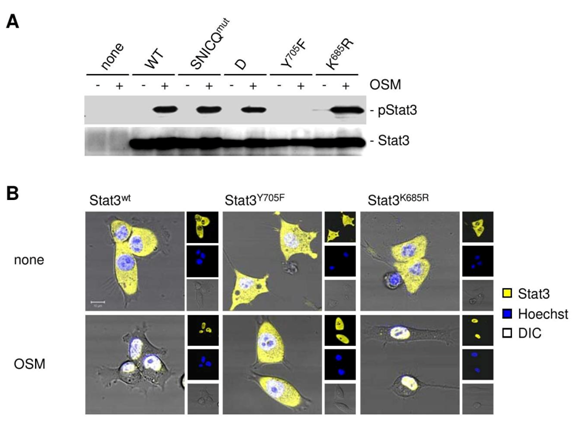
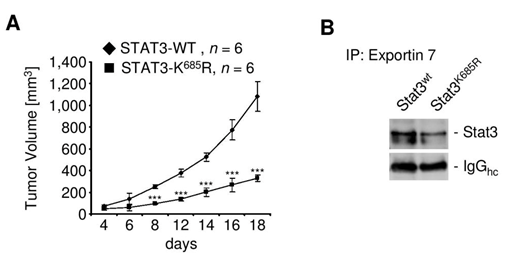
Since exportin 7 was shown to recognize lysine containing motifs in cargo proteins,23 we hypothesized that the main STAT3 acetylation site, lysine K685,24 might be critical for interaction with exportin 7. Acetylation of STAT3 at K685 was readily detectable in human tumor tissues and cell lines (supplementary material (Supplementary Figure S1). Inhibiting STAT3 acetylation using resveratrol, we showed reduced STAT3/exportin 7 interaction with diminished STAT3 acetylation but not phosphorylation (Figure 1D). Furthermore, introducing acetylation mutation in a constitutively activated STAT3 mutant, STAT3C (STAT3C/K685R), resulted in the loss of interaction with exportin 7 (Figure 1E). STAT3 mutation at K685 (STAT3K685R) did not affect STAT3 phosphorylation (supplementary material) (Supplementary Figure S2A) and nuclear translocation upon OSM stimulation (supplementary material) (Supplementary Figure S2B). Cells stably expressing STAT3K685R, form tumors in vivo at significantly reduced rate compared to STAT3wt expressing cells (supplementary material) (Supplementary Figure S3A). Importantly, the reduction of STAT3K685R/exportin 7 interaction was validated using tumor homogenates for co-immunoprecipitation experiments (supplementary material) (Supplementary Figure S3B).
Although exportin 1/Crm1 was suggested potentially being involved in STAT3 nuclear export based on the NES that has a homologue in STAT1, so far no direct evidence prove that is the case. Our results indicate that exportin 7 is the main karyopherin critical for STAT3 nucleocytoplasmic shuttling. Furthermore, we show that STAT3 acetylation site is required for exportin 7-mediated STAT3 nuclear export.
STAT3 lysine acetylation is critical for nuclear retention
To investigate the role of STAT3 acetylation in nuclear retention we assessed STAT3 nuclear egress employing the fluorescence protease protection (FPP) assay21 (Figure 2A). Upon distinct permeabilization of the outer cell membrane but not the nuclear envelope, STAT3wt underwent rapid nuclear egress while STAT3K685R showed an extended nuclear signal (Figure 2B, 2C). Intriguingly, STAT3wt interacted with exportin 7 after 2h upon ectopic stimulation with a STAT3 activator OSM (Figure 2D). Furthermore, RNAi-based silencing of exportin 7 (XPO7) resulted in decreased induction of STAT3 target genes such as VEGF, survivin and BclXL (Figure 2E).
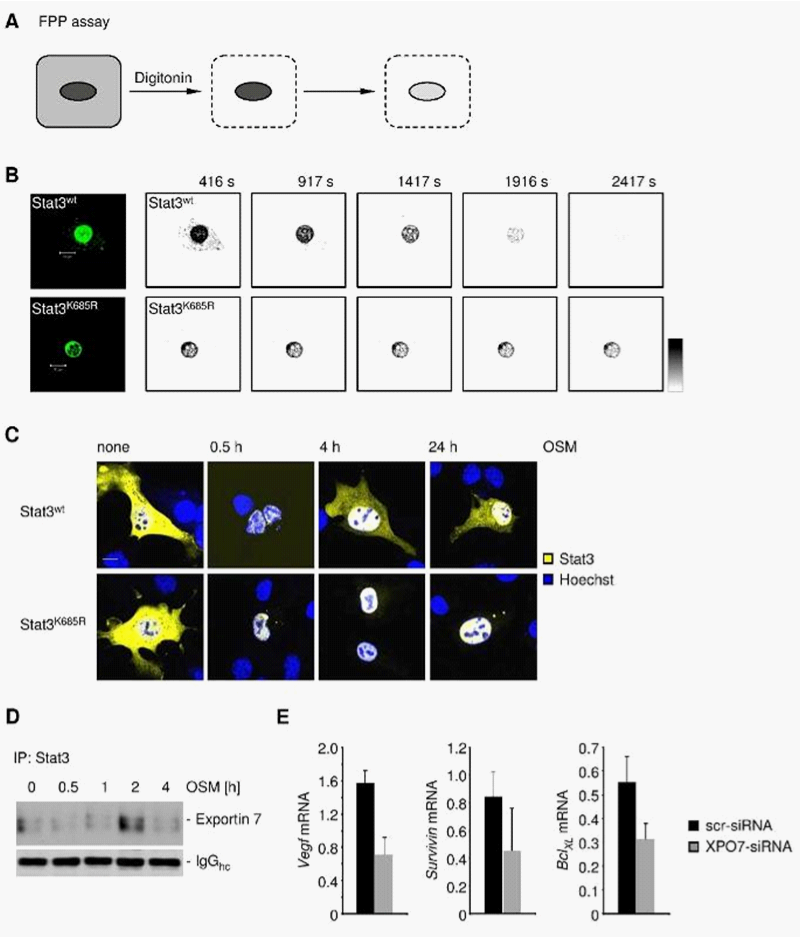
The FPP assay demonstrates that loss of a key acetylation site corrupted efficient STAT3 transport into the cell cytoplasm. The importance of nuclear translocation and export in modulating STAT protein transcriptional activity is well appreciated.7,11,19 Data shown in Figure 2 illustrate the point that loss of STAT3 nuclear egress by silencing exportin 7 can affect STAT3 downstream gene expression.
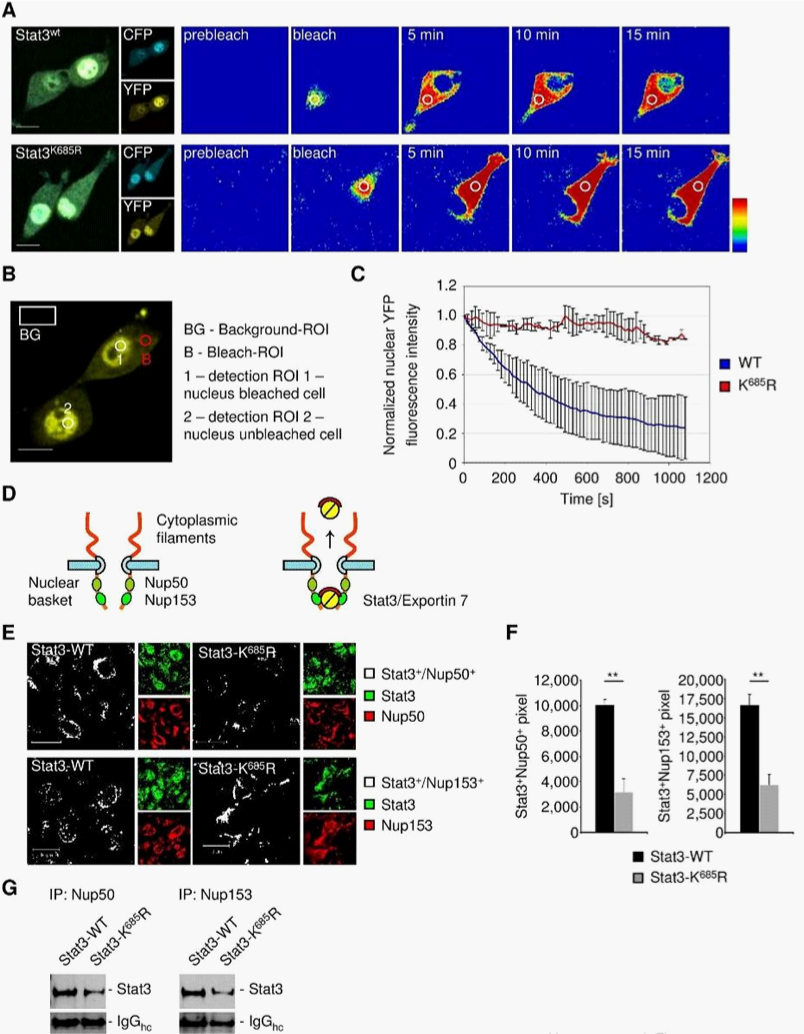
Acetylation is critical for STAT3 nucleocytoplasmic shuttling
To address STAT3 compartmental turnover, we expressed wild-type STAT3 (STAT3wt) or acetylation deficient STAT3 (STAT3K685R) genetically fused to CFP-YFP in 3T3/v-Src cells. The ability of v-Src to activate STAT3 in the 3T3 cells has been well documented.4,25–28 This allowed us to perform intracatenar fluorescence loss after photo-bleaching (iFLAP) experiments in living cells.8,29 As an internal control for visualization of STAT3 protein mobility and spatiotemporal distribution, a neighboring cell was included in the field of view. Cytoplasmic STAT3wt-CFP-YFP or STAT3KR-CFP-YFP was photo-bleached at l=514nm and followed for subcellular distribution by recording CFP and YFP emissions. Consistent with a previous study,8 STAT3wt migrating from the defined cytoplasmic bleach-ROI entered the cell nucleus as fast as 10min. However, nuclear migration of acetylation-deficient STAT3K685R could not be detected within 15min (Figure 3A). Using a more quantitative approach by monitoring nuclear YFP decay under the same experimental setting (Figure 3B), we were able to estimate STAT3 nuclear egress dynamics. While STAT3wt underwent continuous nuclear decay, STAT3K685R fluorescent signals remained constant (Figure 3C).
Nucleoporins of the inner basket of the NPC facilitate the egress of nuclear proteins.16,30,31 Nucleoporins 50 and nucleoporin 153 are major components of the NPC representing a check-point for nuclear exit30,31 (Figure 3D). We therefore analyzed the interaction of STAT3wt and STAT3K685R with nucleoporins to assess nuclear egress efficacy. As shown by confocal microscopy, STAT3wt colocalized with nucleoporin 50 and 153, resulting in nuclear-envelope-like structures (Figure 3E). However, STAT3K685R colocalization with nucleoporins was significantly decreased (Figure 3F), which was concomitant with considerably decreased physical interaction of STAT3K685R with nucleoporins compared to STAT3wt in vivo (Figure 3G) indicating interrupted nucleocytoplasmic shuttling of STAT3K685R.
iFLAP illustrates a requirement of acetylation at lysine K685 of STAT3 for effective nuclear cytoplasmic shuttling. Acetylated STAT3 is also needed for STAT3 interaction with nucleoporins 50 and 153, which form check-points for protein nuclear egress. However, these results do not distinguish whether STAT3 directly or indirectly interact with nucleoporins 50 and 153.
STAT3 acetylation contributes to exportin 7 functional localization
Exportin 7 was initially identified to recognize protein-motifs containing lysine residues. Once exportin 7 engages its cargo, it is thought to exclude cargoes from nuclei, thereby contributing to compartmental distribution of cargo protein.23 Exportin 7 was found in the cell cytoplasm in tumors (Figure 4A, left panel). However, in tumors expressing acetylation-deficient STAT3K685R, exportin 7 was mainly detected in the cell nucleus when compared to STAT3wt expressing tumors (Figure 4A, right panel). Of note, inhibition of STAT3 acetylation did not interfere with exportin 7 mRNA expression levels (Supplementary Figure S4).
To further investigate the role of STAT3 acetylation in subcellular distribution of exportin 7, we engrafted cells stably expressing acetylation-deficient STAT3K685R into athymic mice, followed by intratumoral delivery of a lysine-acetylated peptide spanning STAT3 acetylation site (acet.Stat3 pept.) to mimic STAT3 acetylation activity. As a control, an ovalbumin (OVA) peptide was used. The peptides were delivered using a biolipid-carrier system.32 While exportin 7 was found in the nucleus of tumor cells, as indicated by colocalization of exportin 7 with the nucleic acid stain Hoechst, delivery of acetylated STAT3 peptide resulted in exportin 7 nuclear exclusion and restoration of the functional localization of exportin 7 (Figure 4B), similar to that of STAT3wt expressing tumors (Figure 4A).
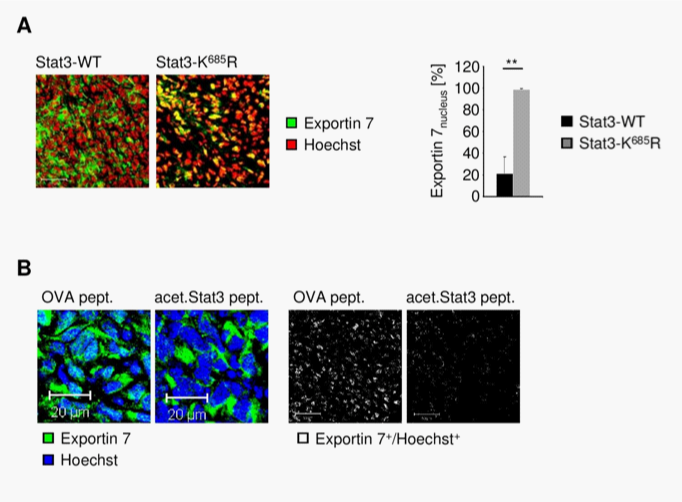
The subcellular localization of exportin 7 in tumor tissues was not defined. Our results indicate that it may be mainly located in the cell cytoplasm. Previous pioneering work has demonstrated that exportin 7 recognizes a broad range of cargoes independent of their specific cellular functions.23 Based on our current study, it appears that STAT3 acetylation is a substrate for exportin 7, contributing to its nucleocytoplasmic shuttling. This finding is consistent with prior studies showing the importance of lysine residues for high affinity binding by exportin 7. The number of proteins actively excluded from nucleus is estimated to be very large.23 The number of proteins potentially acetylated can be very high. However, not all these proteins are regulated by nucleocytoplasmic shuttling. Besides, acetylation site(s) on certain proteins may not be exposed to allow interaction with exportin 7. Perhaps even more important is the fact that acetylation itself can be tightly regulated. In the case of STAT3, acetylation responds to certain cytokines and acetylation of STAT3 is prominent in various human cancers33,34 (Supplementary Figure S1). These findings are also supportive of the notion that STAT3 nuclear export is critical for its reactivation, nuclear translocation and transcriptional activity.
This work is funded in part by R01CA122976. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Andreas Herrmann designed and performed experiments, analyzed data and wrote the manuscript; Sergey Nachaev, Christoph Lahtz, Heehyoung Lee, Anne Schroeder, Brian Armstrong, Claudia Kowolik, Marcin Kortylewski, performed experiments and analyzed data; Richard Jove and Hua Yu wrote the manuscript.

© . This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.