MOJ
eISSN: 2471-139X


Mini Review Volume 9 Issue 1
Laboratorio de Nanobiologia Celular, Departmento de Biologia Celular, Facultad de Ciencias, Universidad Nacional Autonoma de Mexico (UNAM), Circuito Exterior, C.U., 04510, Cd. Mx., Mexico
Correspondence: María de Lourdes Segura-Valdez, Laboratorio de Nanobiologia Celular, Departmento de Biologia Celular, Facultad de Ciencias, Universidad Nacional Autonoma de Mexico (UNAM), Circuito Exterior, C.U., 04510, Cd. Mx., Mexico, Tel +52 55 56 22 49 88
Received: July 15, 2022 | Published: July 20, 2022
Citation: Acosta-Cárdenas J, Jiménez-García LF, Segura-Valdez ML. Speckles in tissues. MOJ Anat Physiol. 2022;9(1):1-3. DOI: 10.15406/mojap.2022.09.00317
In the cell nucleus, splicing factors organize as cell structures called speckles when visualized by fluorescence microscopy. Morphology of speckles is transcription and splicing activity-dependent. While most studies on speckles have been performed using cell lines, results using cells present in tissues are not so abundant. Here we present a minireview on those studies supporting the results available on cells in culture.
Keywords: Cell nucleus, fluorescence microscopy, RNPs, speckles, tissue
IGCs, interchromatin granules clusters; mRNA, messenger RNA; pre-mRNA, pre messenger RNA; RNPs, ribonucleoproteins; PFs, perichromatin fibrils; RNApol II, RNA polymerase II; snRNP, small nuclear ribonucleoprotein
Gene expression, the primary mechanism of regulation of structure and functions in living beings, implies two universal and conserved events (when speaking of the expression that leads to the synthesis of a protein): transcription and translation.1 The main product of transcription is a primary transcript or pre-messenger RNA (pre-mRNA) in need of a maturation process before its export to the cytoplasm. Pre-mRNA is then converted into a mature messenger RNA (mRNA) through co- or post-transcriptional modifications such as capping of the 5´ end, polyadenylation of the 3´end, and removing the introns and joining together the resulting exons by splicing.2 Splicing is driven by a protein-directed metalloribozyme: the spliceosome.3-5 This molecular complex contains ribonucleoproteins and non-ribonucleoproteins splicing factors. The fluorescence immunolocalization of those splicing factors creates a speckled pattern in the nucleoplasm which is composed of 30-50 speckles of irregular shape with diffuse connections among them when observed in culture cells (Figure 1).6 The use of transmission electron microscope techniques has allowed defining that the speckles correspond to the Interchromatinian Granules Clusters (IGC) which are involved in the storage, assembly, and recycling of the spliceosome components. On the other hand, the diffuse areas correspond to the Perichromatin Fibers (PF), and they represent sites of transcription.7-9 Together, the IGC and PF constitute the Splicing Factors Compartments, which are one of the membrane-less compartments that contribute to the compartmentalization of the cellular nucleus.8,10

Figure 1 Speckles in culture cells. Speckled pattern in cultured human fibroblasts observed by immunofluorescence using an antibody against splicing factors. a) Nuclei are labelled as a speckled pattern. b) High magnification of a cell in a). N, nucleus; n, nucleolus. Speckles (large arrows) are embedded in a more diffuse signal (small arrows).
The splicing factors stored in the IGC are recruited to the PF when the transcription by RNA polymerase II (RNA pol II) is activated. In the PF the spliceosome is assembled since splicing is a co-transcriptional event.11-13 Because of the recruitment of the splicing factors from the storage sites (IGCs) to the active transcription sites (PFs), a change in the morphology of the speckles from a rounded and compact pattern to a more diffuse one2 takes place. It has also been proposed that speckles are formed with the participation of specific proteins as SON, in processes involving liquid-liquid phase transitions due to the vicinity of speckles to sites of transcription. Also, the amount of factors in speckles may have to do with the release of splicing factors from transcription sites.14-18
Most studies on speckles have been performed using cell lines in culture. This probably happens due to the easiness of manipulation of the cellular conditions in a cell culture compared to a living animal. Nevertheless, there have been some studies carried out on animal tissues (Figures 2 & 3). Those studies confirmed the presence of a speckled pattern and analyze some of their characteristics and dynamic properties in the studied tissues. For example, George-Tellez and colleagues (2002) confirmed the presence of speckles in several rat tissues as hepatic, pancreatic, uterine, and various types of epithelia as intestinal, uterine, and bronchial. They use immunofluorescence and confocal microscopy to describe the variations in the number and shape of the speckles in the studied tissues which are subject to different transcription programs. Besides, they reported the effects of transcription on the morphology of speckles in a single tissue under distinct stimuli. Specifically, levels of estradiol change during the estrus cycle, and the same happens to the transcriptional activity in reproductive tissues such as the uterus. They found that high hormone concentration in blood (during the proestrus) is related to a pattern of irregular-shaped speckles. While low hormone concentration (during diestrus I) corresponds to a more spherical-shaped one. These results were mimicked experimentally by analyzing the speckle pattern in uterine cells after castration and injection of estradiol. They obtained the same type of results, i. e.: round speckles in the absence of estradiol and irregular speckles when estradiol was present.19
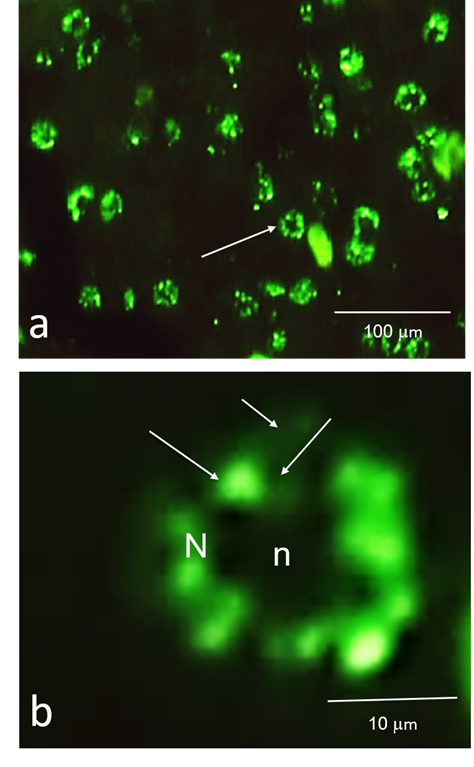
Figure 2 Speckles in tissue. Speckled pattern in lizard (Sceloporous sp) liver tissue sections observed by immunofluorescence using an antibody against splicing factors. N, nucleus; n, nucleolus. b) High magnification of a cell in a). N, nucleus; n, nucleolus. Speckles (large arrows) are embedded in a surrounding diffuse signal (small arrows).

Figure 3 Speckles in tissue. Speckled pattern in rat endometrial epithelial tissue cells observed by immunofluorescence using an antibody against splicing factors. N, nucleus; n, nucleolus. b) High magnification of a cell in a). N, nucleus; n, nucleolus. Speckles (large arrows) are embedded in a surrounding diffuse signal (small arrows).
Mammalian tissues are not the only ones where the speckles have been found. In 2007, Segura-Valdez and colleagues confirmed the presence of a speckled pattern in several tissues of different vertebrate species (chicken, lizard, frogs, fish, and lamprey) and the hemichordate Balanoglossus. In this study, an immunolocalization was performed using as a target a splicing factor (antibody 3C5) known to form part of the speckles in cells grown in culture. Although, the speckles found in this species have not been ultrastructurally characterized.20 In addition, interchromatin granules were isolated from mouse liver tissue to perform proteomic studies, although no fluorescence microscopy was performed,21,22 indicating the presence of a speckled pattern. Moreover, there are some papers also reporting the presence of speckles in plant tissue.23-25
On the other hand, in 1991, Wu and colleagues described in the germinal vesicle of Xenopus a structure that they called snurposomes (from an acronym for snRNP). They are rich in snRNPs and other splicing factors and may function as a site of assembly, storage, and recycling for these factors, but also, they may act as a reservoir of these factors for the embryo. The authors suggest that the snurposomes may be functionally equivalent to the speckles, but given their spherical shape and the diversity found in a single germinal vesicle, it is not accurate to homologize them.26
Speckles have been described in somatic and germinal tissues in vertebrates and a hemichordate in addition to the classical studies performed in cells in culture. Their ultrastructure, functions, and dynamic behavior in non-mammalian vertebrates remained open for investigation.
Jeniffer Acosta-Cárdenas is a graduate student of Posgrado en Ciencias Biológicas- UNAM, supported by a scholarship from CONACyT-México.
No financial interest or any conflict of interest exists.

©2022 Acosta-Cárdenas, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.