MOJ
eISSN: 2471-139X


Mini Review Volume 6 Issue 5
Department of Cell Biology and Anatomy, Rosalind Franklin University of Medicine and Science, USA
Correspondence: Michael P Sarras Jr, Department of Cell Biology and Anatomy, Chicago Medical School, Rosalind Franklin University of Medicine and Science, 3333 Green Bay Road, North Chicago, IL 66044, USA, Tel 520-404-6327
Received: September 11, 2019 | Published: September 16, 2019
Citation: Michael PS. Hydra as a unique model for the study of regenerative mechanisms in metazoans. MOJ Anat & Physiol. 2019;6(5):151-153. DOI: 10.15406/mojap.2019.06.00264
The study of regeneration has led to remarkable contributions to medicine with the advent of stem cell biology and the application of stem cells in the treatment of human trauma and disease. Along with a number of other animals, Hydra represents a premier organism for the study of stem cells and regeneration in general. Initiated by the studies of Trembley in the 1700 hundreds, Hydra has allowed for the basis of biochemical, cellular, mechanical, and molecular studies on the underlying mechanisms of regeneration. By using new molecular techniques one is able to take advantage of the fact that Hydra is under constant renewal and this allows one to thereby dissect details of the renewal process as will be discussed in this review.
Keywords: hydra, regeneration, mechanical mechanisms, cellular mechanisms, molecular mechanisms
Scientific study of regeneration as an aspect of development began in the third century BC with Aristotle who studied development of the chick embryo with a focus on the anatomical origin of organs such as the cardiovascular system.1 Regeneration as a separate area of study began in earnest with Trembley2 who focused his experiments on the freshwater invertebrate, Hydra. Trembley's approach involved experiments in which he would wound adult polyps or graft a segment of Hydra (e.g. the head pole) to adult polyps and observe the subsequent regenerative process the occurred. The strength of Hydra for such studies is its high regenerative capacity, simplified body structure, and state of constant renewal in which all cells of the organism are in constant turnover. Since Trembley, additional organisms have been incorporated as models for the study of regeneration to include such organisms as: planaria, the red spotted newt, the axolotl, and most recently the freshwater fish, zebrafish (Danio renio).3, 4 As stated though, Hydra's is unique among these models because of its high regenerative capacity, simplified body structure, and state of constant renewal. Because of these attributes, studies have been able to target the complete lineage of the epithelial and stem cell populations using advanced techniques as will be discussed.
As indicated, Hydra's strength as a model system for regeneration arises from its 1) simplified structure coupled with its 2) high regenerative capacity. The simplified structure of Hydra is shown in the figure below. It is essential an epithelial organism composed of an ectoderm and endoderm with an intervening extracellular matrix (ECM), termed the mesoglea. Non-epithelial cells reside in both the ectoderm and endoderm. The non-epithelial cells include stem cells that develop into the other cells within the two epithelial layers. An overall diagram of Hydra is shown in Figure 1.
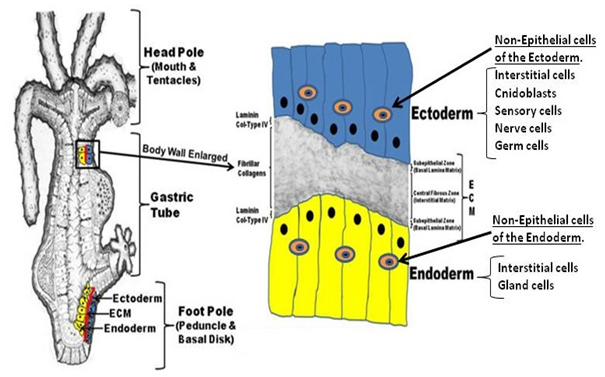
Figure 1 Hydra is a bipolar organism with a body lining composed of an ectoderm and endoderm. Between these layers is an extracellular matrix composed of classical components such as laminin and collagen. Between the ectoderm and endoderm cells reside stem cells called Interstitial cells that lead to the differentiated cells shown in the right column of the figure. It should be noted, that the ectoderm and endoderm epithelial cells are under constant division for renewal of the body lining.
The regenerative ability of Hydra arises mainly from the epithelial cells in interaction with the ECM. This is based on experiments in which all non-epithelial cells are depleted from the organism. In such "epithelial Hydra" the body structure is fully capable of complete regeneration.5 The regenerative capacity of Hydra is so great that pellets of dissociated epithelial cells from adult polyps are capable of reorganizing into the adult polyp structure.6,7
Studies indicate that the mechanisms of Hydra regeneration are based on interactions at multiple organizational levels to include: molecular, cellular, and tissue levels.8 Molecular levels appear to involve 1) head to tail and 2) tail to head gradient systems that help to define polarity of the regenerating organism if the head or foot pool is removed. These lead to the cellular and tissue level interactions that include cell-cell and cell-ECM interactions.4,9–12
There are many approaches to the study of regeneration in Hydra, depending on the type of regeneration being studied. For the purposes of this review we will focus on head regeneration in which the head pole of the adult polyp is surgically removed (by simple ablation) or a second head is transplanted to an intact Hydra and the process analyzed as to what signaling systems are employed by the organism. Based on comparison to other metazoans, Hydra utilizes organizer centers for the definition of axes. At the head pole it has been proposed that a "head organizer" exists that defines the head pole along the body axis.10–12 It is proposed that a head activation (HA) and head inhibition (HI) gradient emanates from this organizer 10. It is proposed that the head organizer is similar to an embryonic organizer. Transplantation of a second head pole to a polyp results in the formation of a second axis being created. Studies indicate that the HA and HI are greatest at the head pole, but the HA is greater than the HI gradient at the head pole. The half life of these gradients is different between HA and HI with HI having a lesser half life. When the head is removed, the HI levels drop and this leaves higher levels of HA which initiates the head regeneration process. While the components of the gradients are still being elucidated, the head organizer appears to be based on such signaling systems as the canonical Wnt pathway.10 This pathway involves genes such as TCF, beta-catenin, and Frizzled to name but a few. A diagram of the Wnt pathway is shown below with all its components (Figure 2). Unique to Hydra are the Wnt-related genes as shown in the Table 1 below.
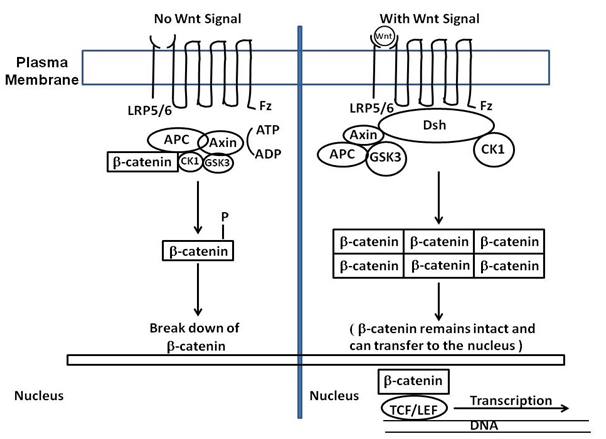
Figure 2 The canonical Wnt pathway (or Wnt/β-catenin pathway) causes an accumulation of β-catenin in the cytoplasm and its eventual translocation into the nucleus to act as a transcriptional co-activator of transcription factors belonging to the TCF/LEF family. β-catenin would not accumulate in the cytoplasm without Wnt, because a degradation complex would degrade it. The degradation complex includes: Axin, adenomatosis polyposis coli (APC), protein phosphatase 2A (PP2A), glycogen synthase kinase 3 (GSK3) and casein kinase 1α (CK1α). The complex degrades β-catenin by targeting it for ubiquitination, which then sends it to the proteasome for digestion. By Wnt binding to Fz (Fizzled) and LRP5/6, the degradation process is blocked. This is due to Wnt causing the translocation of the negative Wnt regulator, Axin, and the destruction complex to the plasma membrane. Phosphorylation by other proteins in the destruction complex subsequently binds Axin to the cytoplasmic tail of LRP5/6. Axin becomes de-phosphorylated and its stability and levels decrease. Dsh then becomes activated via phosphorylation and inhibits the GSK3 activity of the destruction complex. This allows β-catenin to accumulate and localize to the nucleus thereby inducing a cellular response via gene transduction alongside the TCF/LEF (T-cell factor/lymphoid enhancing factor) transcription factors.
Hydra genes of the Wnt family based on genomic studies |
hywnt1 |
hywnt5a |
hywnt8 |
hywnt7 |
hywnt9/10a |
hywnt9/10b |
hywnt9/10c |
hywnt11 |
hywnt16 |
Table 1 A list of Wnt-related genes identified by the Hydra genome projects
These systems have been expanded upon due to the Hydra genome project as well as Hydra transcriptome studies.13,14 These gradient systems also play a role in the position at which buds form along the adult body column.10
As an example of the unique contributions made through the study of Hydra, recent studies have been able to elucidate the genetic pathways that underlie the homeostatic tissue maintenance of the organism.15 These studies sequenced 24,985 Hydra single-cell transcriptomes and generated maps of the Hydra cell states and differentiation molecular sequences for all Hydra cells (to include Hydra's stem cells). The study provided the transcription factors expressed at key differentiation decision steps and by doing so, demonstrated the unique power of Hydra in the elucidation of complex regenerative and developmental processes.
As will be further discussed in a follow-up review, molecular studies (genomic, transcriptome, and cloning) have further expanded our understanding of the gradient signaling system in Hydra. For example, transcriptome studies have identified 10,597 novel Hydra transcripts that encode some proposed 529 conserved proteins. This includes the FGF and TOR signaling systems. Further analysis is underway to further elucidate those systems that form the basis of head regeneration in Hydra and thus far; these studies indicate that signaling systems are highly conserved in metazoans.14

©2019 Michael. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.