MOJ
eISSN: 2471-139X


Mini Review Volume 7 Issue 1
Department of Cell Biology and Anatomy, Chicago Medical School, Rosalind Franklin University of Medicine and Science, USA
Correspondence: Michael P Sarras Jr., Dept. of Cell Biology and Anatomy, Rosalind Franklin University of Medicine and Science, 3333 Green Bay Road, North Chicago, IL 66044, USA, Tel 520-404-6327
Received: January 19, 2020 | Published: January 28, 2020
Citation: Michael PS. Hydra, as a Rosetta stone for deciphering the underlying mechanisms of modern day regenerative medicine. MOJ Anat and Physiol. 2020;7(1):8-10. DOI: 10.15406/mojap.2020.04.00280
A growing and important category of modern medicine is the field of regenerative medicine because through it exists the potential for organ replacement and improved tissue repair procedures. The challenges to regenerative medicine are many because of the complexity of the regenerative process. One approach to deciphering the mechanisms underlying regeneration is analysis of more simplified tissue systems that mimic that seen among all metazoans. To that end many laboratories across the world have focused on organisms such as Hydra (phylum Cnidaria and class Hydrozoa) because its body wall is reduced to an epithelial bilayer with an intervening extracellular matrix (ECM). That fact coupled with its high regenerative capacity (complete body form regeneration from pellets of dissociated epithelial cells isolated from the adult polyp) makes Hydra a powerful tool to investigate the underlying mechanisms of regeneration. Over the years it has become apparent that Hydra and higher vertebrates such as human share a common "developmental tool kit" that is used in their regenerative processes. This review will discuss the 1) cell-cell and cell-ECM processes as well as advances in 2) genomic and transcriptome studies that highlight key aspects of hydra regeneration that can be transferred to the field of regenerative medicine. In this regard, it can be concluded that understanding of basic regenerative mechanisms of Hydra can be applied and used to advance the field of regenerative medicine.
Keywords: regenerative medicine, regeneration, Hydra
ECM, extracellular matrix; Wnt, int/Wingless family of growth factors (to include the: canonical Wnt pathway, the noncanonical planar cell polarity pathway, and the noncanonical Wnt/calcium pathway); Col-Type IV, collagen type IV; RTK, receptor tyrosine kinase; FGF, fibroblast growth factor; BMP, bone morphogenetic protein; TGF, transforming growth factor (sometimes referred to as tumor growth factor)
Modern regenerative medicine can look to the groundbreaking work of Abraham Trembley in the 1700s1–3 as an early foundation for the basis of its understanding of the regenerative process as it applies to the repair of organ dysfunction. The strength of Hydra as regenerative model stems from its simplified body wall structure that is reduced to an epithelial bilayer composed of an ectoderm and endoderm with an intervening extracellular matrix (ECM), termed the mesoglea. The overall structure of hydra is shown in Figure 1.
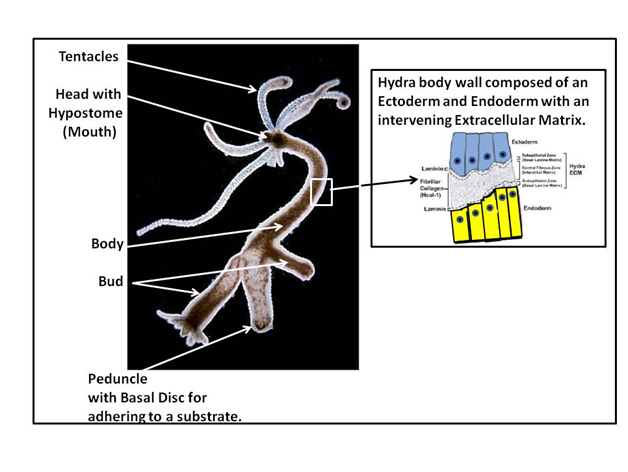
Figure 1 As shown to the left side of Figure 1, Hydra, as a fresh water Cnidarian that is organized as a tubular organism with a gastric cavity. Morphologically it is structured with a head and tail pole. This involves a 1) head with a mouth (hypostome) and associated tentacles, 2) a body with outgrowing buds for asexual reproduction of the organism (hydra also utilized sexual reproduction through egg and sperm fusion), and 3) a foot pole with its peduncle and basal disc. The basal disc produces glycoproteins that allow the organism to attach to any substrate. The image to the right depicts the body wall structure of Hydra, with its ectoderm (blue) and endoderm (yellow) and its intervening extracellular matrix (ECM) composed of components reflective of modern day ECM (e.g. Laminins and collagens as well as many other ECM molecules).4–10
Hydra (phylum Cnidaria and class Hydrozoa) arose early during metazoan evolution (approximately 580 million years ago before the divergence of the protosomes and deuterostomes groups). Hydra is the first of existing organism with defined epithelial cell layers (seen as a bilayer) with junctional complexes present at the apical pole of the cells. The epithelial cells of Hydra continuously turn over as seen in human skin epithelium. Hydra therefore has true epithelial tissues as compared to the more primitive groups such as the sponges. In this regard, sponges are thought to have diverged before Hydra. To appreciate the powerful extent of Hydra regenerative capacity it should be pointed out besides its ability to regenerate both its head and foot pole after excision of these poles from the body, Hydra also has the ability to regenerate its entire adult epithelial structure to include its ECM from pellets of cells obtained from non-enzymatically dissociated adult Hydra polyps.11 This includes polyps devoid of any interstitial cells (termed "epithelial hydra").
Hydra also has interstitial cells that reside within the ectoderm and endoderm layers. These interstitial cells include 1, nematocytes (the stinging cells from which Cnidaria derives its name) 2) nerve cells, 3) and a number of other cell types to include i-cells that are stem cells from which the other interstitial cells continuously arise. It should be noted however, that the high regenerative capacity of Hydra is solely due to the epithelial cells because polyps lacking any i-cells are fully capable of complete body regeneration (they are unable to feed themselves; however, because of the lack of nerve cells).12,13 Because of its simplified structure and its high regenerative capacity, Hydra has been used as a model for analysis regenerative mechanisms that underlie our understanding of the regenerative mechanisms of human tissues and organs. Hydra is pertinent to our understanding vertebrate regenerative mechanism because its molecular structural components (cellular and ECM) mimic those seen in higher vertebrates to include humans.4–10
This review will discuss Hydra regeneration as a basis for our understanding of regenerative medicine in terms of its 1) cell-cell and cell-ECM interactions and its 2) molecular interactions based on current advances in genomic and transcriptome studies.
Cellular and mechanical processes pertaining to hydra regeneration
Gradient systems: Based on the pioneering studies by developmental biologists such as Lewis Wolpert and Hans Bode, we have a firm understanding of the basic tenants of gradient systems in Hydra.13,14 From a morphogenesis standpoint early studies established that if an adult hydra polyp is cut in-half, the upper head half will regenerate a new basal disc, and the lower basal disc half will regenerate a new head. This occurs no matter where along the body one makes the incision. Based on grafting experiments, it was deduced that hydra polarity is dictated by a series of morphogenetic gradients that permit the head and basal disc to only form at specific locations along the longitudinal body axis. Additional grafting experiments defined the existence of a head activator gradient (highest at the hypostome) and a basal activator gradient (highest at the basal disc). Further studies defined 1) head activator gradient and 2) a head inhibitor gradient as well as a 3) basal disc activator gradient and 4) basal disc inhibitor gradient. These inhibitor and activator gradients also provide instructions as to which end is apical and which end is basal. Therefore, when the head is removed, the head inhibitor is no longer is made, and this results in the head activator to induce a new head. Accordingly, the region with greatest amount of head activator will form a head structure; thereby restoring a gradient equilibrium from head to foot.
Cell-cell and Cell-ECM interactions: Cell-Cell and Cell-ECM interactions are fundamental to development processes and are an integral aspect to Hydra regeneration.9,15,16 In regard to cell-cell interactions, Hydra has a broad range of adhesion molecules and completes observed in higher vertebrates and humans such as Integrins, tight junctional complexes (septate junctions, in Hydra), and gap junctions; to name a few.17,18 These molecules function not only in adhesion and signal transduction, but also serve to facilitate cell trans-differentiation processes that are essential for regeneration to occur. More recent studies has shown that the epithelial cytoskeleton facilities the regenerative process.19 These cell-cell interactions interplay with cell-ECM interactions to add another level of control in the regenerative process. As previously published by our laboratory, ECM formation is tightly coupled with the regenerative process as reviewed by Sarras.6,7,20–22 The laboratory of Sarras has established also established that hydra's ECM components mimics that seen in higher vertebrates and man.7 Briefly, our studies have shown that within one hour of surgical decapitation, the cut ends of the head pole fuse and the elastic properties of the ECM result in it being retracted so that the newly sealed epithelium lacks an intervening matrix. The epithelium acquires a flattened morphology due to the lack of an intervening ECM. By three hours after the sealed epithelium bilayer forms, an up-regulation of ECM components occurs within the epithelial cells. By seven hours of this process, basal lamina components (laminin and Col-TypeIV) are translated and secreted between the epithelial bilayer and this is accompanied by the epithelial bilayer taking on its normal cellular morphology. Following the appearance of the basal lamina, and twenty hours after decapitation was initiated, interstitial fibrillar collagens begin to be translated and appear between the two previously formed basal lamina layers adjacent to the basal extracellular border of the ectoderm and endoderm.20 This is likely the result of ECM-epithelial signals due because it can be perturbed by blocking antibodies to hydra laminin. Once the interstitial matrix is formed, the ECM is now completely polymerized and a normal adult Hydra body wall structure is reformed. Normal head regeneration then is observed. These studies are reinforced by subsequent studies that show the same type of cell-ECM interactions occur when the ECM component blocking experiments are repeated using the hydra-cell-pellet system11 where pellets are formed from dissociated hydra cells and these pellets then go on to form intact adult polyps.6,11
Genomic and transcriptome processes pertaining to Hydra regeneration
The application of genomic and transcriptome studies has greatly advanced our understanding of the underlying molecular mechanisms of hydra regeneration23,24 and this has implications to higher vertebrates and man. The publicly available Hydra and Hydractinia genome portals (https://research.nhgri.nih.gov/hydra/; https://research.nhgri.nih.gov/hydractinia), are constantly being expanded and thereby increasing our understanding of the genomic aspects of hydra regeneration. Additionally, single-cell RNA-seq for transriptonic analysis is emerging as a powerful tool to identify rare cell types and transient cell types that occur during regeneration. For example, studies at UC at Davis have initiated studies from 25,000 single cells of Hydra and have been able to describe the differentiation pathways from the tree lineages of adult polyp.25 Their data indicates that neurons and gland cells share a common progenitor cell during the normal turnover of cells in the adult Hydra. A combination of genomic and transriptonic studies indicate common molecular pathways shared by Cnidarians and higher vertebrates such as the 1) Wnt pathway, 2) RTK pathway with shared ligands such as FGF, 3) BMP pathway with the multiple Smads (transcription factors), 4) TGF pathways, and 5) Notch pathway. Additionally, epigenetic mechanisms are also functional in Hydra as seen in higher vertebrates and in human diseases such as Diabetes mellitus.26 These genomic and transcriptome studies reinforce the highly conserved nature of regenerative processes among metazoans from Hydra to Homo sapiens.
From the early studies of Trembley in the 1700s to current genomic and transcriptome studies, Hydra has provided a window into the underlying mechanisms of regeneration. Based on 1) the shared "regenerative tool kit" of molecules observed among metazoans, 2) the simplified structure of Hydra, and 3) its high regenerative capacity; we have been able to elucidate regenerative mechanisms that can then be translated to and studied in higher vertebrates such as Homo sapiens. Although much has been learned, it is apparent that we have learned only the most rudimentary aspects of a highly complicated process. The application of what we learn from Hydra will further our understanding of regeneration and allow us to apply these mechanisms to regenerative medicine which is the key to organ replacement and tissue repair.
The author wishes to express his appreciation to the National Institutes of Health, USA (DK092721) for funds that supported preparation and writing of this review.
The author has no conflict of interests or commercial interests as related to the information provided in this review.
None.

©2020 Michael. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
This is a modal window.