Journal of
eISSN: 2475-5540


Research Article Volume 1 Issue 7
1Department of Cancer Biology and Pharmacology, University of Illinois College of Medicine, USA
2Department of Neurosurgery, University of Illinois College of Medicine, USA
3Department of Neurology, University of Illinois College of Medicine, USA
4Illinois Neurological Institute, USA
Correspondence: Krishna Kumar Veeravalli, Department of Cancer Biology and Pharmacology, University of Illinois College of Medicine at Peoria, 1 Illini Dr. Peoria, IL, USA, Tel (309) 671-8442
Received: December 23, 2016 | Published: December 30, 2016
Citation: Chelluboina B, Nalamolu KR, Klopfenstein JD, et al. Stem cell treatment after ischemic stroke alters the expression of dna damage signaling molecules. J Stem Cell Res Ther. 2016;1(7):281-288. DOI: 10.15406/jsrt.2016.01.00049
Accumulating evidence suggests that oxidative DNA damage plays a critical role in cell death associated with ischemic stroke. Endogenous oxidative DNA damage can be detected in the ischemic brain during the stages that precedes the manifestation of cell death and is believed to trigger cell death via various intracellular signaling pathways. Inhibiting the signaling associated with DNA damage induction or facilitating the signaling associated with the DNA repair process can be neuroprotective against brain injury after ischemic stroke. Recent reports demonstrated that human umbilical cord blood-derived mesenchymal stem cells (HUCB-MSCs) prevented the upregulation of apoptotic signaling pathway molecules and thereby attenuated the extent of apoptosis after focal cerebral ischemia as well as improved the neurological recovery. Therefore, we hypothesized that HUCB-MSCs treatment after focal cerebral ischemia prevents the overexpression of molecules associated with DNA damage induction as well as augments the expression of molecules associated with DNA repair process. In order to test our hypothesis, we administered HUCB-MSCs (0.25x106cells/animal) intravenously via tail vein to male Sprague-Dawley rats that were subjected to a two-hour middle cerebral artery occlusion followed by one-day reperfusion. Ischemic brain tissues obtained from various groups seven days’ post reperfusion were subjected to DNA damage signaling pathway PCR microarray. Our results demonstrated the induction of both DNA damage inducing and repair genes after focal cerebral ischemia and reperfusion. HUCB-MSCs treatment downregulated the DNA damage inducing genes and upregulated the DNA repair genes without disturbing the endogenous defense mechanisms.
Keywords: ischemic stroke, stem cells, dna damage, repair, upregulation, downregulation
Ischemic stroke is one of the leading causes of death worldwide and is the biggest reason for serious long-term disability. Accumulating evidence suggests that oxidative DNA damage plays a critical role in cell death associated with ischemic stroke. Endogenous oxidative DNA damage can be detected in the ischemic brain during the stages that precede the manifestation of cell death, and is believed to trigger cell death via various intracellular signaling pathways, including apoptosis. DNA repair is the endogenous defense mechanism to combat oxidative DNA damage. The capacity for DNA repair may influence the pathological outcome of ischemic stroke. Inhibiting the signaling associated with DNA damage induction or facilitating the signaling associated with DNA repair process can be neuroprotective against brain injury after ischemic stroke.
Stem cell transplantation offers a promising therapeutic strategy to treat ischemic stroke. In addition to preventing the ongoing damage, stem cell transplantation actually repairs the injured brain. It has emerged as a potential regenerative treatment to reduce post-stroke handicap. Human umbilical cord blood-derived mesenchymal stem cells (HUCB-MSCs) possess several advantages over other types of stem cells including those derived from bone marrow.1 They also possess multipotent differentiation potential and thus can be induced to differentiate into cells of multiple lineages such as adipocytes, osteocytes, chondrocytes, myocytes, hepatocytes, neurons, astrocytes and oligodendrocytes.1-8 Treatment with HUCB-MSCs significantly reduced the infarct size, improved survival and offered neurological recovery in animals subjected to ischemic stroke.9-11 In addition, HUCB-MSCs treatment after focal cerebral ischemia prevented the upregulation of apoptotic signaling pathway molecules and thereby attenuated the extent of apoptosis.12,13
Based on the reported data and our preliminary investigations, we hypothesize that HUCB-MSCs treatment after focal cerebral ischemia prevents the overexpression of molecules associated with DNA damage induction as well as augments the expression of molecules associated with the DNA repair process. In order to address our hypothesis, we administered HUCB-MSCs to rats that were subjected to a two-hour middle cerebral artery occlusion (MCAO) followed by one-day reperfusion. To our knowledge, this is the first study to explore the effect of HUCB-MSCs treatment on the expression of DNA damage signaling pathway molecules.
Animals
Adult male Sprague-Dawley rats weighing 220-250g were procured from Envigo laboratories (USA). Animals were housed in a 12-h light/dark cycle with controlled temperature and humidity and free access to food and water. Animals were randomly assigned to five groups. Group 1 was used as sham. Group 2 animals were subjected to a two-hour MCAO procedure without any further treatment and sacrificed on seventh day post-MCAO. Groups 3 and 4 animals were treated with 0.25x106 HUCB-MSCs immediately after reperfusion through intravenous and intra-arterial (via internal carotid artery) routes, respectively, and sacrificed one hour after stem cell administration. Group 5 animals were intravenously administered (via tail vein) 0.25x106 HUCB-MSCs one-day after reperfusion and sacrificed on seventh day post-MCAO. The Institutional Animal Care and Use Committee (IACUC) of the University of Illinois College of Medicine at Peoria approved all surgical interventions and post-operative animal care. All animal experiments were conducted in accordance with the IACUC guidelines. In addition, all the procedures that were performed on the animals were in compliance with the approved IACUC protocol.
Experimental MCAO model
After the rats reached a weight of 260±5 g, they were subjected to right MCAO procedure as previously described.13 Briefly, a ventral midline incision (~25mm) was made in the neck and the right common carotid, internal carotid, and external carotid arteries were surgically exposed. The external carotid artery was permanently ligated rostral with one ligature. A micro-aneurysm clip was applied to the external carotid artery near its bifurcation with the internal carotid artery. A small puncture opening was made in the external carotid artery. The monofilament was inserted through the opening, and the other loose ligature was tightened around the lumen containing the monofilament. The micro-aneurysm clip was removed from the external carotid artery, and the monofilament was then gently advanced from the lumen of the external carotid artery into the internal carotid artery for a distance of ~19 to 20 mm beyond the bifurcation of the common carotid artery. The skin on the neck incision was closed with surgical wound clips. To restore the blood flow 2 hours after MCA occlusion, the surgical site was re-opened by removing the wound clips. The knot was loosened, the monofilament was withdrawn, and the knot was re-tied to stop bleeding. The skin was sutured to close the neck incision. After MCAO procedure, all animals were treated with appropriate analgesics and antibiotics and subjected to the approved post-procedural care.
Stem cell treatment
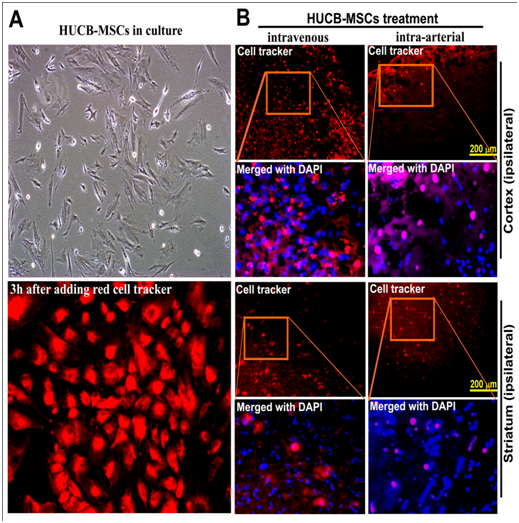
Cryo-preserved HUCB-MSCs obtained from Vitro Biopharma (Golden, CO) were used to establish cultures in MSC-GRO low serum complete MSC medium according to the provided instructions. Cultures were maintained at 37˚C in a humidified atmosphere containing 5% CO2 with a change of culture medium twice a week. When the cell cultures were about 80% to 90% confluent, cells were split and sub-cultured. Cells were detached, washed twice with sterile PBS, counted and suspended in sterile saline prior to intravenous administration. In order to demonstrate the entry of HUCB-MSCs into the ischemic brain tissue after intravenous and intra-arterial administrations, HUCB-MSCs (0.25x106/animal) were incubated in culture with red-cell tracker (Invitrogen, Oregon, USA) for 3 hours before administration to groups 3 and 4 rats. Group 5 animals subjected to the MCAO procedure were intravenously injected with 0.25x106 HUCB-MSCs via tail vein, 24 hours post-MCAO procedure and sacrificed on the seventh day.
Brain tissue sectioning and staining
One hour after HUCB-MSCs administration, rats from groups 3 and 4 were placed under deep anesthesia with pentobarbital and perfused through the left ventricle with 70-100 mL of PBS, followed by 100-150 mL of 10% buffered formalin (Fisher Scientific, NJ). The brains were then removed and embedded in Tissue-Tek® O.C.T. 4583 embedding medium. Serial coronal brain sections of the frozen brain tissue were cut at a thickness of 20-30μm using a cryostat. Brain sections were stained with DAPI, cover slipped, and observed using a confocal microscope (Olympus Fluoview).
RNA extraction and cDNA synthesis
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) from the ischemic ipsilateral brain regions of rats sacrificed on the seventh day post-MCAO procedure. Briefly, total RNA was extracted with TRIzol and precipitated with isopropyl alcohol, washed in ethanol, and resuspended in RNase-free water. One μg of total RNA was reverse transcribed into first-strand cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Indianapolis, IN) according to the manufacturer’s instructions. The cDNAs obtained from this process were used for rat DNA damage signaling RT2 Profiler PCR array.
DNA damage signaling PCR array
We used the RT2 Profiler Rat DNA damage signaling PCR Array (SA Biosciences). Each array contains a panel of 96 primer sets of 84 relevant DNA damage signaling pathway focused genes plus five housekeeping genes and three RNA and PCR quality controls. The cDNAs obtained from the brain sections of sham, untreated and HUCB-MSCs-treated rats sacrificed on day seven post-MCAO procedure were loaded onto microarray plates along with FastStart SYBR Green (Roche, Indianapolis, IN) according to the manufacturer’s instructions. The sample-loaded microarray plate was subjected to real-time PCR analysis in iCycler IQ (Multi Color Real-Time PCR Detection System, Bio-Rad Laboratories, Hercules, CA). Real-time PCR was carried out under the following conditions: one cycle of 95oC for 10 minutes, and 40 cycles of 95˚C for 15 sec and 60oC for 1 minute. We performed the PCR microarray analysis on the samples obtained from three different animals per group.
Stem cells migrated to the ischemic brain after administration
Presence of red fluorescence in the cortex and striatal regions of the ischemic hemisphere indicated that the administered HUCB-MSCs (0.25x106/animal) reached their site of action as expected (Figure 1). In addition to their administration by intravenous route via tail vein, HUCB-MSCs incubated with red cell tracker were also administered one hour after reperfusion subsequent to a two-hour ischemia to another group of animals by intra-arterial route via internal carotid artery. Administration of HUCB-MSCs by either of these routes resulted in the migration of these cells to the rat ischemic brain regions. Therefore, we selected the intravenous route to administer HUCB-MSCs for further experiments in this study. In order for the HUCB-MSCs to migrate to the ischemic brain after their administration, they should be permeable through the blood brain barrier (BBB). Recently, we reported the possible disruption of BBB, which occurred as early as two-hours post MCAO without reperfusion in the same rodent model of focal cerebral ischemia.14 Therefore, it is quite possible for the HUCB-MSCs administered one hour after reperfusion subsequent to a two-hour ischemia, to reach various regions of the ischemic brain.
Ischemia and reperfusion leads to the overexpression of DNA damage signaling molecules
PCR microarray of DNA damage signaling pathway molecules performed on the ischemic brain samples obtained from ischemia (two-hour)- and reperfusion (seven days)-induced rats revealed overexpression of the several molecules tested (Figure 2). The overexpressed molecules include those involved in the DNA damage induction, DNA repair, apoptosis and cell cycle (Table 1). Prominently upregulated genes of the DNA damage signaling pathway after cerebral ischemia and reperfusion include Atm (~181 fold vs. sham), and Brca1 (~165 fold vs. sham). Rad51c is the only gene that is several fold downregulated (~105 fold vs. sham) after cerebral ischemia and reperfusion.
Gene |
Gene Description |
Fold Regulation |
Upregulated genes |
||
Abl1 |
C-abl oncogene 1, receptor tyrosine kinase |
2.7 |
Apex1 |
APEX nuclease (multifunctional DNA repair enzyme) 1 |
2.3 |
Atm |
Ataxia telangiectasia mutated homolog (human) |
181.7 |
Atrx |
Alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) |
3.3 |
Bard1 |
BRCA1 associated RING domain 1 |
2.9 |
Bbc3 |
Bcl-2 binding component 3 |
2.3 |
Blm |
Bloom syndrome, RecQ helicase-like |
2.8 |
Brca1 |
Breast cancer 1 |
164.9 |
Brca2 |
Breast cancer 2 |
4.8 |
Cdc25a |
Cell division cycle 25 homolog A (S. pombe) |
2.4 |
Cdkn1a |
Cyclin-dependent kinase inhibitor 1A |
2.2 |
Chek2 |
CHK2 checkpoint homolog (S. pombe) |
2.1 |
Csnk2a2 |
Casein kinase 2, alpha prime polypeptide |
3.9 |
Ddb2 |
Damage specific DNA binding protein 2 |
5.1 |
Ddit3 |
DNA-damage inducible transcript 3 |
14.9 |
Ercc1 |
Excision repair cross-complementing rodent repair deficiency, complementation group 1 |
2.6 |
Fanca |
Fanconi anemia, complementation group A |
2.7 |
Fancc |
Fanconi anemia, complementation group C |
4.1 |
Fancd2 |
Fanconi anemia, complementation group D2 |
2.9 |
Fen1 |
Flap structure-specific endonuclease 1 |
2.7 |
Lig1 |
Ligase I, DNA, ATP-dependent |
2 |
Mbd4 |
Methyl-CpG binding domain protein 4 |
2.2 |
Mlh1 |
MutL homolog 1 (E. coli) |
4 |
Mlh3 |
MutL homolog 3 (E. coli) |
2.5 |
Mpg |
N-methylpurine-DNA glycosylase |
2 |
Mre11a |
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
3.3 |
Msh2 |
MutS homolog 2 (E. coli) |
3.3 |
Parp2 |
Poly (ADP-ribose) polymerase 2 |
2.9 |
Pcna |
Proliferating cell nuclear antigen |
2.8 |
Pms1 |
Postmeiotic segregation increased 1 (S. cerevisiae) |
2.1 |
Pole |
Polymerase (DNA directed), epsilon |
4.2 |
Polh |
Polymerase (DNA directed), eta |
2.1 |
Poli |
Polymerase (DNA directed), iota |
2.3 |
Ppm1d |
Protein phosphatase 1D magnesium-dependent, delta isoform |
2 |
Prkdc |
Protein kinase, DNA activated, catalytic polypeptide |
2.3 |
Pttg1 |
Pituitary tumor-transforming 1 |
2.9 |
Rad17 |
RAD17 homolog (S. pombe) |
2.1 |
Rad18 |
RAD18 homolog (S. cerevisiae) |
2.5 |
Rad51b |
RAD51 paralog B |
2.6 |
Rad52 |
RAD52 homolog (S. cerevisiae) |
2.2 |
Rad9 |
RAD9 homolog (S. pombe) |
3.8 |
Rpa1 |
Replication protein A1 |
2.4 |
Sirt1 |
Sirtuin (silent mating type information regulation 2 homolog) 1 (S. cerevisiae) |
2.9 |
Smc1a |
Structural maintenance of chromosomes 1A |
4.6 |
Sumo1 |
SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) |
14.3 |
Tp53bp1 |
Tumor protein p53 binding protein 1 |
2.5 |
Ung |
Uracil-DNA glycosylase |
3.2 |
Xpc |
Xeroderma pigmentosum, complementation group C |
2.8 |
Xrcc2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
2.1 |
Xrcc6 |
X-ray repair complementing defective repair in Chinese hamster cells 6 |
2.6 |
Downregulated genes |
||
Pms2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
-3.3 |
Rad51c |
Rad51 homolog c (S. cerevisiae) |
-105 |
Xrcc1 |
X-ray repair complementing defective repair in Chinese hamster cells 1 |
-2.2 |
Table 1 Regulation of DNA damage signaling pathway molecules after focal cerebral ischemia and reperfusion in rats
Injury = 2-hour ischemia + 7days reperfusion
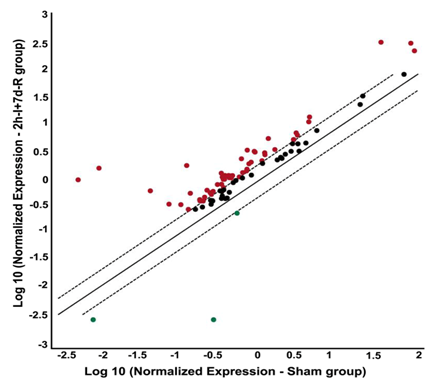
Figure 2 Focal cerebral ischemia followed by reperfusion alters the gene expression profile of DNA damage signalling pathway molecules. PCR microarray analysis of rat DNA damage signalling pathway genes from the cDNAs obtained from the untreated MCAO-subjected, and sham operated rats. Scatter plot shows the fold difference in the expression of each gene in untreated MCAO subjected versus sham operated rat brain samples (from ipsilateral region). Red dots indicate the upregulated gene expression and blue dots indicate the downregulated gene expression.
n=3; I: Ischemia; R: Reperfusion
Stem cell treatment prevented the upregulation of DNA damage signaling molecules
The overexpression of DNA damage signaling pathway genes was prevented in stroke-induced rats that were subjected to HUCB-MSCs treatment. This effect of HUCB-MSCs treatment was clearly evident from downregulated gene expression, especially when the gene expression after HUCB-MSCs treatment was compared with the gene expression in untreated stroke-induced rats (Figure 3). The only genes that were prominently upregulated after HUCB-MSCs treatment as compared to untreated, stroke-induced rats include Rad51c (~71 fold) and PMS2 (~63 fold) (Table 2). Prominently downregulated genes after HUCB-MSCs treatment as compared to untreated, stroke-induced rats include Lig1 (~151 fold), Mgmt (~1211 fold), Rad17 (~139 fold), Rad18 (~208 fold), and Smc1a (~1523 fold).
Gene |
Gene Description |
Fold Regulation |
Upregulated genes |
||
Pms2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
63.4 |
Rad51c |
Rad51 homolog c (S. cerevisiae) |
71.4 |
Downregulated genes |
||
Abl1 |
C-abl oncogene 1, receptor tyrosine kinase |
-4.2 |
Apex1 |
APEX nuclease (multifunctional DNA repair enzyme) 1 |
-5.4 |
Atm |
Ataxia telangiectasia mutated homolog (human) |
-4.4 |
Atrx |
Alpha thalassemia/mental retardation syndrome X-linked (RAD54 homolog, S. cerevisiae) |
-2 |
Bax |
Bcl2-associated X protein |
-2.1 |
Bbc3 |
Bcl-2 binding component 3 |
-10.9 |
Blm |
Bloom syndrome, RecQ helicase-like |
-3.6 |
Brca1 |
Breast cancer 1 |
-3.9 |
Brca2 |
Breast cancer 2 |
-5.3 |
Cdc25a |
Cell division cycle 25 homolog A (S. pombe) |
-3.8 |
Cdc25c |
Cell division cycle 25 homolog C (S. pombe) |
-2.4 |
Cdkn1a |
Cyclin-dependent kinase inhibitor 1A |
-3.3 |
Csnk2a2 |
Casein kinase 2, alpha prime polypeptide |
-6.1 |
Ddb2 |
Damage specific DNA binding protein 2 |
-11.3 |
Ercc1 |
Excision repair cross-complementing rodent repair deficiency, complementation group 1 |
-4.7 |
Ercc2 |
Excision repair cross-complementing rodent repair deficiency, complementation group 2 |
-2.4 |
Exo1 |
Exonuclease 1 |
-2.7 |
Fanca |
Fanconi anemia, complementation group A |
-8.9 |
Fancc |
Fanconi anemia, complementation group C |
-3.9 |
Fancd2 |
Fanconi anemia, complementation group D2 |
-3.6 |
Fancg |
Fanconi anemia, complementation group G |
-2.2 |
Fen1 |
Flap structure-specific endonuclease 1 |
-4.6 |
Gadd45a |
Growth arrest and DNA-damage-inducible, alpha |
-2.4 |
Gadd45g |
Growth arrest and DNA-damage-inducible, gamma |
-3.4 |
Hus1 |
HUS1 checkpoint homolog (S. pombe) |
-3.5 |
Lig1 |
Ligase I, DNA, ATP-dependent |
-151.4 |
Mbd4 |
Methyl-CpG binding domain protein 4 |
-4.3 |
Mgmt |
O-6-methylguanine-DNA methyltransferase |
-1211.6 |
Mlh1 |
MutL homolog 1 (E. coli) |
-5.2 |
Mlh3 |
MutL homolog 3 (E. coli) |
-3.5 |
Mpg |
N-methylpurine-DNA glycosylase |
-2.3 |
Mre11a |
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
-5.7 |
Msh2 |
MutS homolog 2 (E. coli) |
-4.2 |
Nbn |
Nibrin |
-3.2 |
Nthl1 |
Nth (endonuclease III)-like 1 (E.coli) |
-4.2 |
Ogg1 |
8-oxoguanine DNA glycosylase |
-9.4 |
Parp1 |
Poly (ADP-ribose) polymerase 1 |
-91.3 |
Parp2 |
Poly (ADP-ribose) polymerase 2 |
-14.1 |
Pcna |
Proliferating cell nuclear antigen |
-3.8 |
Pms1 |
Postmeiotic segregation increased 1 (S. cerevisiae) |
-3.2 |
Pole |
Polymerase (DNA directed), epsilon |
-5.3 |
Polh |
Polymerase (DNA directed), eta |
-5.5 |
Poli |
Polymerase (DNA directed), iota |
-4.6 |
Ppm1d |
Protein phosphatase 1D magnesium-dependent, delta isoform |
-2.9 |
Prkdc |
Protein kinase, DNA activated, catalytic polypeptide |
-3.9 |
Pttg1 |
Pituitary tumor-transforming 1 |
-4.8 |
Rad1 |
RAD1 homolog (S. pombe) |
-11.5 |
Rad17 |
RAD17 homolog (S. pombe) |
-139.4 |
Rad18 |
RAD18 homolog (S. cerevisiae) |
-208.3 |
Rad50 |
RAD50 homolog (S. cerevisiae) |
-4.4 |
Rad51b |
RAD51 paralog B |
-3.8 |
Rad52 |
RAD52 homolog (S. cerevisiae) |
-4.3 |
Rad9 |
RAD9 homolog (S. pombe) |
-6.8 |
Rnf8 |
Ring finger protein 8 |
-3.3 |
Rpa1 |
Replication protein A1 |
-5.8 |
Sirt1 |
Sirtuin (silent mating type information regulation 2 homolog) 1 (S. cerevisiae) |
-3.2 |
Smc1a |
Structural maintenance of chromosomes 1A |
-1523 |
Smc3 |
Structural maintenance of chromosomes 3 |
-2.2 |
Sumo1 |
SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) |
-11.6 |
Topbp1 |
Topoisomerase (DNA) II binding protein 1 |
-2.6 |
Tp53 |
Tumor protein p53 |
-2 |
Tp53bp1 |
Tumor protein p53 binding protein 1 |
-5.4 |
Ung |
Uracil-DNA glycosylase |
-2.9 |
Xpc |
Xeroderma pigmentosum, complementation group C |
-4.8 |
Xrcc2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
-2.8 |
Xrcc6 |
X-ray repair complementing defective repair in Chinese hamster cells 6 |
-2.9 |
Table 2 Regulation of DNA damage signaling pathway molecules after stem cell therapy in stroke-induced rats
Injury = 2-hour ischemia + 7days reperfusion
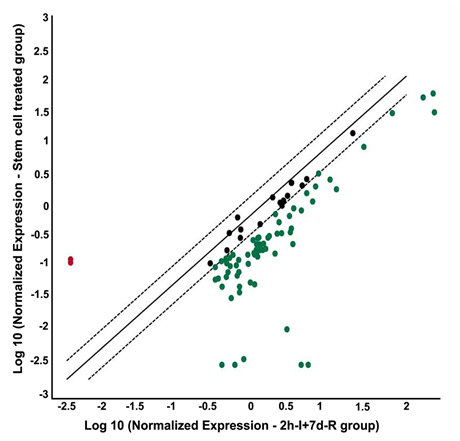
Figure 3 Stem cell treatment after focal cerebral ischemia alters the gene expression profile of DNA damage signalling pathway molecules. PCR microarray analysis of rat DNA damage signalling pathway genes from the cDNAs obtained from the ischemic region of HUCB-MSCs treated (0.25x106 cells/animal, intravenous), and untreated MCAO-subjected rats. Scatter plot shows the fold difference in the expression of each gene in HUCB-MSCs treated versus untreated MCAO subjected rat ischemic brain samples. Red dots indicate the upregulated gene expression and blue dots indicate the downregulated gene expression.
n=3; I: Ischemia; R: Reperfusion
In the current study, focal cerebral ischemia and reperfusion in a rat model upregulated the gene expression of several DNA damage signaling pathway molecules as expected. These overexpressed genes include those that are associated with DNA damage induction, DNA repair, apoptosis and cell cycle. The overexpression of genes associated with the DNA repair activity could be attributed to the body’s defense mechanism in response to the ongoing brain damage after ischemic stroke. Interestingly, the expression of both Atm, and Brca1, the genes associated with DNA repair, was several fold higher than any other genes tested including the DNA damage inducing genes. This data clearly demonstrates how aggressive the endogenous defense mechanisms in response to injury are.
The downstream relaying of DNA damage, mainly double-strand breaks, involves the activation of Atm. Activation of Atm could phosphorylate various protein targets to activate cell cycle check-points and repair the DNA damage.15 If the damage cannot be repaired, Atm directs the cell to apoptosis. Whether the outcome is DNA repair or cell death likely depends on both the extent and the duration of injury. The increased mRNA expression of Atm after ischemic stroke in the current study could be the body’s defense mechanism for DNA repair against the ongoing DNA damage in the ischemic brain. However, we do not know whether all the increased Atm expression in this case is leading to the repair of DNA damage or apoptosis. We recently reported that the HUCB-MSCs treatment after ischemic stroke inhibited apoptosis and the extent of ischemic brain damage.12 HUCB-MSCs treatment in the present study prevented Atm overexpression only to some extent (~4 fold down regulation vs. untreated stroke-induced group) indicating that the overexpression of Atm after ischemic stroke is mostly associated with DNA repair. Brca1 is one of the downstream targets of Atm. Brca1 is a required cofactor for ATM-catalyzed phosphorylation of several targets including p53 and Chk2.16 After phosphorylation by Atm, H2AX recruits repair proteins such as Brca1 to the sites of DNA damage. Similar to their effect on Atm expression, HUCB-MSCs treatment in the present study prevented Brca1 overexpression only to some extent (~4 fold down regulation vs. untreated stroke-induced group) indicating that the overexpression of Brca1 is required for DNA repair. In contrast to Atm and Brca1 expression, Rad51c is the only gene that is several fold downregulated after cerebral ischemia and reperfusion. Rad51c gene is a member of the Rad51 family. Rad51 family members are known to be involved in the homologous recombination and repair of DNA. Although HUCB-MSCs treatment prevented the downregulation of Rad51c, the reason for the downregulation of Rad51c after ischemic stroke is unclear, and needs further investigation.
Prominent upregulation of Pms2 was noticed in HUCB-MSCs treated rats when compared to untreated, stroke-induced rats. The Pms2 gene is a member of a set of genes known as the mismatch repair genes. It provides instructions for making a protein that plays an essential role in DNA repair. This protein helps fix mistakes that are made when DNA is copied (DNA replication) in preparation for cell division. The Pms2 protein joins with another protein called Mlh1 to form a protein complex, which coordinates the activities of other proteins that repair mistakes made during DNA replication.
As expected, HUCB-MSCs treatment prominently reduced the gene expression of Parp1, as compared to untreated, stroke-induced rats. Parp1 plays a detrimental role in cell death process after ischemic brain injury. It serves as a sensor to bind DNA strand breaks, especially single-strand breaks and activate downstream signaling events. Evidence suggest that post-ischemic activation of Parp1 occurs in practically every cell type of the affected brain region and significantly contributes to the extent of final damage.15 Translocation of apoptosis inducing factor (AIF) from mitochondria to the nucleus is a key step in the Parp1 cell death process.17,18 Parp1 inhibition attenuated the migration of AIF from mitochondria to the nucleus and protected neurons from death after oxygen–glucose deprivation in vitro and focal cerebral ischemia in vivo.19 Further, our recent study demonstrated that the HUCB-MSCs treatment after ischemic stroke prevented the overexpression of AIF protein and the extent of apoptosis.12 The results of the present study suggest that the prevention of AIF overexpression and a reduction in the extent of cell death after HUCB-MSCs treatment in stroke-induced animals, as reported earlier, could have been mediated through the downregulation of Parp1.
Several fold downregulation in gene expression of Lig1, Mgmt, Rad17, Rad18, and Smc1a was noticed in HUCB-MSCs treated rats when compared to untreated, stroke-induced rats. Lig1, also called DNA ligase 1, encodes a member of the ATP-dependent DNA ligase protein family. The encoded protein functions in DNA replication, recombination, and the base excision repair process. Mgmt repairs the naturally occurring mutagenic DNA lesion and prevents mismatch and errors during DNA replication and transcription. The protein encoded by Rad17 gene is required for cell cycle arrest and DNA damage repair in response to DNA damage. Rad18 is another DNA damage repair gene. The protein encoded by the Smc1a gene interacts with Brca1 and is phosphorylated by Atm, indicating a potential role for this gene in DNA repair. The downregulation of these DNA damage repair genes could be the consequence of reduced DNA damage associated with HUCB-MSCs treatment.
Altogether, our results demonstrate that the increased expression of DNA damage repair genes such as Atm and Brca1 after ischemic stroke could be the result of endogenous defense mechanisms in response to the DNA damage. Our results also demonstrate that the HUCB-MSCs treatment after ischemic stroke down regulates DNA damage inducing genes such as Parp1 and upregulates the DNA damage repair genes such as Pms2 without disturbing the endogenous defense mechanisms.
We thank Illinois Neurological Institute, OSF Health Care System for the financial assistance. We also thank Amy Gries and Christina Constantinidou for manuscript preparation and review.
The author declares no conflict of interest.

©2016 Chelluboina, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.