Journal of
eISSN: 2475-5540


Short Communication Volume 8 Issue 1
Immunology of Invertebrates, Orléans University, France
Correspondence: Michel Leclerc, Immunology of Invertebrates, Orléans University, France, 556 rue Isabelle Romée, 45640 SANDILLON
Received: May 24, 2023 | Published: May 30, 2023
Citation: Leclerc M. Other sea star Igkappa gene cloning assay in E. coli with new parameters. J Stem Cell Res Ther. 2023;8(1):25-26 DOI: 10.15406/jsrt.2023.08.00159
The plasmid vector pET-28b(+) named “Young” was produced according a work of 2014.1 This construct is designed to allow the expression of a 13.6 kDA protein with a C-terminal 6histag. It is supposed to be an anti-HRP (Horse-radish peroxydase protein). This protein was not expressed in first E. coli: we attempt to explain this phenomenon.
In a first time we recall the construction of our vector with the help of Clinisciences (Paris, France)
The first step was the de novo gene synthesis of the sequence below.
tGACTGCTGCTATGCGTGGCAACATGGCGTCTCTATGGATGTTCTTCTTTGTCgTGGGG
ATAACTTTACAACGGAGT
TTGGCGATTTACACGTTTCGCGAGCAACCGTCGGACACTAGCGCGTTGCAGGGGAGC
ACAGTGGTGCTTCACTGCT
CCGTTGAGCAGTACATAAACACCACGGCCATCGTTTGGTGGAGCCGTGACTCGGTCA
TCAGCCACAACAAAGACCTGAAACTGTCCAGTCTAAACACCGACCAGCTCCAAAGGT
ACTCGATTTCAGGCGACGCATCTCGGGGGGAATTCAACCTTAAAATAGTGAACTTTA
CCGNCACAGACGCCGCCAGTTACCGCTGTCAGATGTTTGCGA
The second step consist of the cloning of the sequence above in pET-28b vector by seamless cloning. Briefly, Seamless cloning was originally described by Daniel G. Gibson,2 of the J. Craig Venter Institute. The exonuclease-based method is performed under isothermal conditions after linear insert and vector are prepared by PCR and/or restriction digestion. Three enzymatic activities are employed: a 5’exonuclease generates terminal cohesive ends (overhangs), a polymerase fills in the gaps of the annealed single-stranded regions, and a DNA ligase seals the nicks. We present now the “schema” of our plasmid with the following abbreviations:
position 12 - 467: f1 bacteriophage origin of replication; arrow indicates direction of (+) strand synthesis
position 560-1375: aminoglycoside phosphor transferase - confers resistance to kanamycin or G418
position 1497-2085: high-copy-number ColE1/pMB1/pBR322/pUC origin of replication
position 2271-2413: basis of mobility region from pBR322
position 2515-2706: Rop protein, which maintains plasmids at low copy number
position 3515-4597: lac repressor
position 4598-4675: lacI promoter
position 4984-5002: promoter for bacteriophage T7 RNA polymerase
position: 5003-5027: lac repressor encoded by lacI binding site
position 5042-5064: efficient ribosome binding site from bacteriophage, T7 gene 10 (Olins and Rangwala, 1989)
position 5071-5430: your gene (M. Leclerc's gene)
position 5341-5448: 6xHis affinity tag
position 5515-5562: transcription terminator for bacteriophage T7 RNA polymerase the vector is 5587 pb in total.
In fact secondly clinisciences with my agreement purposed such a nucleotide sequence and protein one:
Nucleotide sequence
atgcgtggcaacatggcgtctctatggatgttcttctttgtcgtggggataactttacaacggagtttggcgatttacacgtttcgcgagcaaccg
tcggacactagcgcgttgcaggggagcacagtggtgcttcactgctccgttgagcagtacataaacaccacggccatcgtttggtggagccgtgac
tcggtcatcagccacaacaaagacctgaaactgtccagtctaaacaccgaccagctccaaaggtactcgatttcaggcgacgcatctcggggggaa
ttcaaccttaaaatagtgaactttaccgccacagacgccgccagttaccgctgtcagatgtttgcgctcgagcaccaccaccaccaccactga
Protein sequence
MRGNMASLWMFFFVVGITLQRSLAIYTFREQPSDTSALQGSTVVLHCSVEQYINTTAIVWW
SRDSVISHNKDLKLSSLNTDQLQRYSISGDASRGEFNLKIVNFTATDAASYRCQMFALEHHHHHH
Several colonies from the transformation of BL21(DE3) (Novagen) with this construct were tested to find the clone allowing the best expression yield.(collaboration with R D -Biotech (Lyon, France)
Several conditions were tested (agitation, inducing concentration and temperature) and resulting fractions from bacterial lysis were analyzed by SDS-PAGE (Figure 1).
Results are expressed in Figure 1 with different parameters (agitation, inducing concentration and temperature)
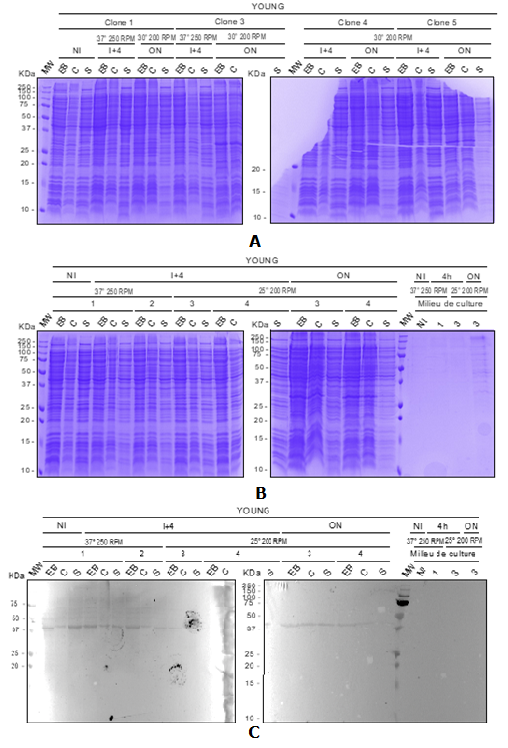
Figure 1 Expression test analysis. SDS-PAGE 15 %, coomassie blue staining A) induction at 37°C 250rpm with 1 mM IPTG; induction at 30°C 200rpm with 1 mM IPTG and B) induction at 37°C 250rpm with 0.5 mM IPTG; induction at 25°C 200rpm with 1mM IPTG; C) Western blot analysis anti Histag.
EB: total lysate; S:Supernatant obtained after EB centrifugation (soluble fraction); C:Pellet obtained after EB centrifugation (insoluble fraction); NI: non induced bacteria; 4: 4H post- induction; O/N:induction Overnight
Despite several expression tests, it was not possible to demonstrate the expression of the protein of interest. Indeed, the theoretical molecular weight of the protein is 13.6 kDa. No overexpression band is observed at this mass. We observe a band of higher intensity at about 25 kDa, however it is not revealed in WB.
Sequence analysis indicates the presence of a signal sequence. The presence of the signal sequence could have caused the production of the protein in the inclusion body. However, this can hardly explain the lack of expression. The pET28 plasmids are conventionally used in the laboratory and it is very rare to have observe no expression even in western blot. It is possible that this is due to the protein itself or the lack of sequence optimization for the bacterial expression system.
So, it seems it was not possible to demonstrate the expression of the protein of interest, may be also we did not use the method of amplification as demonstrated in 2014 with primers.3 These primers acted at the start of the experimentation.
None.
Author have no conflict of interest.

©2023 Leclerc. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.