Journal of
eISSN: 2475-5540


Research Article Volume 2 Issue 4
1Guru Angad Dev Veterinary and Animal Science University, India
2Department of Veterinary Anatomy, Guru Angad Dev Veterinary and Animal Science University, India
Correspondence: Ratan K Choudhary, School of Animal Biotechnology, Guru Angad Dev Veterinary and Animal Science University, Ludhiana - 141004, Punjab, India
Received: February 28, 2017 | Published: April 24, 2017
Citation: Choudhary RK, Choudhary S, Verma R, et al. Mucin 1 aberrantly expresses in goat mammary carcinoma. J Stem Cell Res Ther. 2017;2(4):126-131. DOI: 10.15406/jsrt.2017.02.00072
The objective of this study was to determine if expression of mucin 1 (MUC1) and abundance of collagen fibers is associated with the incidence of goat mammary cancer. MUC1 often expressed in metastatic cancers and has been used as a diagnostic marker for metastatic progression. Similarly, collagen density affects tumor microenvironment and promotes invasion, angiogenesis and migration of cells. Mammary parenchymal tissues were collected from 17 goats from an abattoir and grouped into lactating (n=3), mastitic (n=6) and mammary cancer (n=8). Protein localization of MUC1 and cell proliferation markers were analyzed by immunohistochemistry. Quantification of transcripts of cancer cell markers and cell proliferation markers were evaluated using real time qPCR. Distribution of total collagen was assessed by picrosirius red staining. Goat mammary cancers were evident by the loss of cellular architecture (from mild to severe distortion of cytoarchitecture) of the mammary glands. This study provides evidence of aberrant expression of MUC1 and increased collagen density in goat mammary cancer. In the literature, MUC1-positive cancer cells are aggressive and results in poor prognosis. Presence of high incidence of mammary cancer in goat is alarming, knowing that the occurrence mammary cancer in ruminant is rare.
Keywords: goat mammary carcinoma, collagen fiber, mucin 1, cell proliferation, gene expression
MUC1, mucin1; qPCR, quantitative polymerase chain reaction; ECM, extra-cellular matrix; PSR, picrosirium red; RT-PCR, reverse transcriptase polymerase chain reaction; MEC, mammary epithelial cells; H&E, hematoxylin and eosin; Ki67, marker of proliferation ki-67; FNDC3B, fibronectin type iii domain containing 3b; CCNB, ccyclin B1; MSI1, musashi 1; PCNA, proliferating cell nuclear antigen
Incidence of mammary cancer in ruminant is rare. The exact reasons of low incidence of mammary cancer in ruminant are not known. The reason of low incidence of mammary cancer in ruminant is likely due to high rate of pregnancies.1 Hormones like estrogen, play an important role in hyperplasia and neoplasia of mammary tissues and increased parity shortens exposure of estrogen and thus reduce incidence of cancer in goats. In addition to this, other factors such as vegetarian diet with high fiber content, short lifespan of animals, continuous milking of the glands and other factors like mammary stem cells. A survey of screening cancers in goats conducted in Africa and none of the cancer were identified to be the mammary cancer.2 A number of studies proposed stem cells to be the origin of breast cancer3–5 and these stem cells are dynamic rather than static where progesterone hormone targets mammary stem cells in transforming breast cancer.6 This could be the reason that cancer stem cells share gene signatures with mammary stem cells.7 Incidence of mammary tumors in dogs and cats is very high. In fact, 50% of tumors in dogs are mammary tumors almost three times higher than in woman. The reasons of high incidence of mammary cancer in dogs, cats and human could be dietary habits (meat eater) and high intake of carcinogens through diet as they are higher on food chain.1 Therefore, it deserves attention towards medical perspective as how mammary glands of ruminants (goat) are protected from being cancerous.
Mucin 1 (MUC1) encodes for a membrane-bound protein that has essential role in providing protective mucosal barrier and cell signaling. O-glycosylated mucin protein has two terminals, N-terminal alpha subunit (MUC1-N) and C-terminal beta subunit (MUC1-C), former being involved in cell adhesion and later one involved in cell signaling. MUC1 protein is expressed by luminal epithelial cells of mammary tissue on their apical surfaces and provides protective barriers against pathogens. MUC1- N terminal and MUC1-C terminal forms heterodimer and expresses on apical border of mammary epithelial cells. Due to loss of cell polarity, during transformation of normal cell to cancer cell, MUC1-C monomer is positioned entire over the cell membrane. Thus, overexpression and aberrant localization of MUC1-C has been associated with mammary cancer. In general, more than 90% of the mammary cancers overexpress MUC1 proteins.8 However, heterogeneity of MUC1 expression has been seen in 14 breast carcinoma cell lines.9 Variability in amount of MUC1 in breast cancer ranges from high to low expression. High cytoplasmic expression and aberrant localization of MUC1 was associated with poor prognosis.10 Cellular proliferation activity of cancer depends upon the grade of tumors. Generally, high cell proliferation activity is seen in high-grade tumors like that of MUC1 expression. However, immuno expression of MUC1 is independent of expression of Ki67.11
It has been well documented that extracellular matrix (ECM) is the most abundant component of tumor microenvironment. It provides physical scaffold to bind cells and signaling molecules that affects cell migration, adhesion and proliferation.12 Collagen is the most abundant macromolecules of ECM and naturally occurs in mammary parenchyma. During cancer progression, ECM undergoes constant architectural changes evident by degradation, increased cross-linking and stiffening of collagen. In breast cancer, collagens are thick, linearized and thick to promote metastasis unlike that of healthy breast tissue where collagens are curly and smooth.13 Thus, high collagen density in mammary gland promotes tumor progression.
Tissue samples
Goat mammary samples were collected from local abattoir. Exact age of the animals were not known at the time of tissue collection. Based on mammary gland cyto architecture evaluation,14 tissues were divided to 3 groups namely, lactating (n=3), mastitic (n=6) and cancer (n =8). Histological screening of carcinomas microscopically reviewed by hematoxylin-eosin stained and immuno stained sections followed by panel of markers with their aberrant expression. The microscopic features of lactating, mastitic and cancer tissues were evaluated and discrepancies were resolved by simultaneous reexamination with MUC1 and Ki67 staining. MUC1 is luminal epithelial cell marker and Ki67 is a cell proliferation marker.
No ethical approval was needed for goat mammary tissue samples as these samples were collected from slaughterhouses. However, consent for conducting this piece of research work obtained from Head of the Department of the School of Animal Biotechnology.
Picrosirius red staining of collagen fibers
Picrosirius red (PSR) staining of collagen fibers was done as per the published literature15 with slight modifications. Briefly, 5µm thick sections were deparaffinized and hydrated to water. Nuclear staining was done using Weigert’s haematoxylin for 7min and then washed in the running tap water for 10min followed with 0.1% PSR in saturated picric acid solution for 1h. Stained slides were rinsed twice in acidified water. Finally, sections were dehydrated in absolute alcohol, cleared in xylene and mounted with DPX (Sigma, CA, USA).
Quantification of staining intensity
In order to count PSR stained area of lactating, mastitic and cancer tissue ImageJ (ver 1.51f) software was used. A total of 10-14 representative photomicrographs (at 400 X magnification) were captured in *tiff format from each tissue (one tissue/animal) from epithelial compartment and stromal compartments in focus separately. PSR stained red areas of collagen were selected by placing marks of different colors to generate the immuno histeochmistry index. Pixel intensity of the images calibrated with scale bar thus areas thus integrated density (IntDen) estimated. IntDen of stained areas was assessed by setting a “color threshold” using the thresholding tool. The image was opened and then the following steps applied. (1) Select “Image-Adjust-Color Threshold.” The setting of “Hue, Saturation and Brightness” was adjusted by sliders until all the stained areas are selected (Table 1). (2) After choosing it to “Dark background” for stained area, “Set” was clicked to threshold of the image. (3) Select “Analyze-Set Measurements” and “Area”, “Standard deviation”, “Integrated density”, “Limit to threshold” and “Display label” were selected. Finally, stained and unstained areas were analyzed by selecting (4) “Analyze-Measure” to get the results in tabular form. Results were saved individually for each animal, separately for epithelial and stromal compartments. Data pooled to each category of the animal and analyzed. Comparative analyses performed using the nonparametric Kruskal Wallis test followed by the Mann-Whitney test. The significance was set at P<0.05.
Hue |
Saturation |
Brightness |
||
PSR |
For quantifying lumen area |
Lactating and mastitis: 0-149 |
0-255 |
135-255 |
Cancer: 0-255 |
0-60 |
180-255 |
||
For quantifying collagen area |
Lactating and mastitis: 90-255 |
50-255 |
135-255 |
|
Cancer: 0-255 |
70-255 |
180-255 |
||
MUC1 |
For quantifying MUC1 stained area |
0-255 |
90-255 |
0-255 |
For quantifying MUC1 unstained area |
0-255 |
30-255 |
0-255 |
|
Ki67 |
For quantifying Ki67 stained area |
0-255 |
90-255 |
158-255 |
For quantifying Ki67 unstained area |
0-255 |
30-255 |
0-255 |
|
Table 1 Color thresholds of hue, saturation and brightness for quantification of integrated density collagen area in goat mammary tissues
Immunostaining of MUC1 and Ki67
Immunohistochemical staining was performed according to procedures previously described14 with minor modifications. Briefly, slides were deparaffinized in xylene and antigen was retrieved in Tris-EDTA buffer (homemade, pH 9.0). After rinsing tissue sections with phosphate buffered saline (PBS), slides were dipped in 3% hydrogen peroxide in water for 20min to quench endogenous peroxidase activity and rinsed for 10min with two changes of PBS. Background staining was prevented by incubating the sections with 2.5% ready to use horse serum supplied with secondary antibody for 30min at room temperature. After protein block, slides incubated overnight at 4°C in a moist chamber with ready-to-use primary antibodies of anti-MUC1 and anti-Ki67 (Biogenex, USA). Sections were washed in phosphate buffer saline (3X5min) before application of ready to use peroxidase conjugated secondary antibody (ImmPRESS universal antibody polymer detection kit, Vector Lab, Burlingame, CA, USA). The peroxidase activity sites were visualized using 3,3-diaminobenzidine (ImmPACT DAB/NovaRed Peroxidase (HRP) Substrate kit, Vector lab). Finally, tissue sections rinsed with distilled water, counterstained with Mayer’s hematoxylin. After dehydration, tissues sections cleared in xylene and mounted with DPX. Tissue sections in which primary antibody were omitted, served as negative control.
RNA isolation and PCNA transcript quantification
Around ten mg of frozen tissue stored in RNAlater (Ambion) was thawed and lysed in 1ml of Trizol (Ambion) using handheld tissue homogenizer (Cole-Parmer, IL, USA). Lyzed samples were incubated for 5min at room temperature (RT) and then centrifuged at 12000xg for 5min at 4°C to remove remaining tissue debris, if any. Supernatants transferred to another fresh 2ml nuclease free tube. A 200µl of chilled chloroform was added, mixed vigorously for 20sec, and incubated at RT for 15min. Mixture centrifuged for 15min to remove aqueous solution. From this onwards, GenElute mammalian total RNA isolation kit (Sigma-Aldrich, CA, USA) protocol was followed. Briefly, aqueous solution containing total RNA was transferred to GenElute filtration column (blue color column) fitted with 2.0ml collection tube, to shear big sized DNA and centrifuged for 1min at 12000xg at 4°C. An equal volume of 100% ethanol added to the filtrate in collecting tube and mixed thoroughly with 1ml micropipette. After quick but proper mixing, samples were loaded into GenElute nucleic acid binding column (red color column). Upon brief centrifugation for 30 sec, flow through was discarded and binding columns were washed with 250µl of wash solution1. DNA was digested using On-Column DNase I Digestion Set (DNASE70; Sigma-Aldrich). After incubation with DNase1 for 15min at RT, columns were washed with 250µl of wash solution 1 and centrifuged at maximum speed for 15sec. Binding column was transferred to new 2ml collection tube washed with 500µl of wash solution 2. Third column washed with 500µl of wash solution 2 and centrifuged for 2min at maximum speed. Upon discarding flow through, an additional centrifugation step carried for 1min at maximum speed to remove residual wash solution 2. Finally, RNA was eluted from filtration column in 40µl of elution buffer.
RT-PCR and quantitative PCR
Reverse transcription of total RNA was performed using iScript™ cDNA Synthesis Kit (BioRad, CA, USA). Briefly, 1µg of total RNA were incubated with 4µl of iScript master mix (5X), 1µl iScript RT and volume were made upto 20µl with nuclease free water. Reaction mixtures were incubated for 5min at 25°C followed by 30 min at 42°C. RT reaction stopped by incubating mixtures for 5min at 85°C. The cDNA thus made, diluted to 1:10 and 3µl of diluted cDNA and used as template for quantification of gene transcripts. RPS23 gene used as endogenous control for the relative quantification of gene transcripts. Primer sequences, annealing temperature and their product sizes are given (Table 2). Two-step qPCR amplification was preceded by sample initial denaturation for 3min at 95°C in thermal cycler (CFX-96, BioRad) using SYBR Green (iQ™, SYBR® Green super mix, BioRad, CA, USA). Each cycle, of 40 cycles, contain denaturation period for 30sec at 98°C, annealing of primers for 30sec at 56 or 60°C, depending upon the target. A melt curve analysis of amplified product was performed to check specificity of the amplified product. Delta delta Ct (∆∆Ct) method was used to quantify expression of genes (log 2 fold change) in mammary cancer samples in comparison to lactating tissues.16
S. No. |
Gene Name |
Gene Function |
Primer Pairs (5’------- 3’) |
Tm |
Product size |
1 |
FNDC3B |
Cancer cell marker, Bovine mammary stem cell Marker |
F: ACCACCTACAAGACCCCTCA |
56 |
150 |
2 |
MUC1 |
Luminal epithelial cell marker and marker of cell differentation |
F: ATTGCCCTGGTTGTGTGTCA |
56 |
111 |
3 |
CCNB1 |
Makes cell enter into G1 to M phase |
F: TCATGCAGGATACCTATGTGCC |
56 |
107 |
4 |
MSI1 |
Mammary stem cell marker |
F: ATGATGCCATGCTGATGTTTG |
56 |
119 |
5 |
PCNA |
Cell proliferation marker |
F: ATCAGCTCAAGTGGCGTGAA |
60 |
142 |
6 |
RPS23 |
A ribosomal protein, Reference gene for mammary tissue |
F: CCCAATGATGGTTGCTTGAA |
60 |
101 |
Table 2 List of qPCR primers, annealing temperatures and their product sizes
Gross morphological examination of mammary tissue followed by histomorphology of H&E stained sections revealed three types of mammary tissues- lactating, mastitic and cancer. Samples of mammary cancer showed variation in tissue organization. Out of 8 samples, 6 were lactating and 2 were prepubertal animals. In one type of goat cancer, tissue was well-defined ductal structures and epithelial cell proliferation towards the ductal lumen (Figure 1A & 1B). While in another type of cancer, there was disruption of alveolar structure (Figure 1C & 1D). In lactating mammary cancer, alveolar structure could be traced with the help of staining of collagen fibers.

Figure 1 Types of mammary cancers of prepubertal and lactating goats. Panel A-B: Inward growth of MEC with intact cytoarchitecture of goat mammary carcinoma. Panel C-D: Complete loss of cytoarchitecture of goat mammary carcinomas. Magnification 400 X
Quantitation of collagen fibers
To understand the collagen density in mammary tissue of lactating, mastitic and cancerous glands, we estimated pixel intensities of collagen fiber present in epithelial and stromal compartments of mammary tissues. In lactating glands, thin rims of collagen fibers were present around the alveoli in the basement membrane (Figure 2). Thickness of collagen fiber varied depending upon the size of the alveoli. A more distended alveolus had thinner rims of collagen fiber. Fibers were loose, curly and thin in epithelial compartment. Stromal compartment collagen fibers were occupied fully except the lumen of blood vessels and empty spaces (Figure 2). In cancer tissue, collagen fibers were more straight, rigid and thick (Figure 2). In metastasis, straight and rigid collagen fibers promote invasion and migration of cancer cells.12
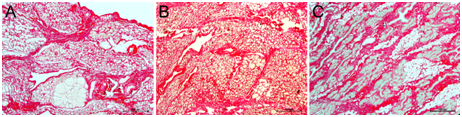
Figure 2 Picrosirius red (PSR) staining of lactating (A), mastitic (B) and cancer of goat mammary glands. Magnifications 400 X.
Density of collagen fibers in lactating mammary gland (Figure 3A & 3B) was lower than that of collagen fibers present surrounding the tumor cells (Figure 3C & 3D). Surface plot of collagen fibers mastitic and cancer tissues is shown in pixel density of region of interests. In mastitis, density of collagen fiber was increased due to reduced surface area of secretory alveoli. Rims of alveoli were thick and collagen deposition was in homogenous fashion. Interestingly, scanty amount of collagen was noticed on the surface of inflammatory cells (polymorphonuclear cells) released into alveolar lumen (arrow; Figure 3E) indicating role of collagen in regulating mastitis related immune infiltration. These mastitic tissue samples were collected from slaughterhouse and hence more likely to be chronic in nature as they were naturally infected rather than experimentally induced mastitis. A Kruskal-Wallis H test showed that there was not a significant difference in stromal collagen content (based on integrated pixel density of area occupied by collagen fibers) between lactating, mastitis and mammary cancer (p=0.3), with a mean rank intensity score of 32.41 for lactating, 37.77 for mastitis and 29.35 for cancer tissue. However, there was a significant difference (p=0.05) in overall epithelial collagen content between lactating, mastitic and cancer, with a mean rank intensity score of 42.35 for lactating, 42.59 for mastitic and 30.17 for cancer tissue. Mann-Whitney U test revealed significant (p=0.03) difference between mastitic and mammary cancer tissue collagen in epithelial compartment (Figure 3F).
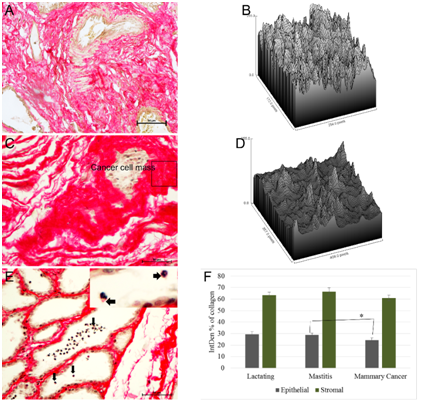
Figure 3 Quantification of PSR staining. PSR quantified in epithelial and stromal compartments of lactating, mastitic and cancer tissues. Integrated density (IntDen) of collagen fiber (expressed in terms of intensity of collagen fiber, of total image intensity) in lactating gland (A) was measured. Surface plot of collagen fiber ROI from lactating tissue (B). In cancer (C), collagen fiber was densely packed around cancer cell mass as show in surface plot of ROI (rectangle area) (D). In mastitic tissue (E) a pinch of collagen fiber (arrows and inset picture) was associated with inflammatory cells present in alveolar lumen. Semi-automatic quantification of PSR stained sections revealed difference in collagen area of mastitic and cancer tissue in stromal compartment (F)..
Immunolocalization of MUC1 and Ki67
Pattern of MUC1 expression was cytoplasm of the alveolar cells located towards apical side (Figure 4A). In mastitis, expression of MUC1 was cytoplasmic but not typically towards apical side of alveolar cells, but diffused in whole cytoplasm (Figure 4B). In some cases, reduced expression of MUC1 expression was noticed. Reduction of MUC1 expression indicated reduction of functional differentiation of mammary epithelial cells (MEC). Histomorphological features of poorly differentiated MEC can be observed by visualizing cellular morphology wherein the cells are smaller in size, absence of fat vacuole inside the cytoplasm and reduced cytoplasmic area.17,18 In mammary carcinomas, aberrant expression of MUC1 was seen. MUC1 was located in the nuclei plus cytoplasm of majority of the MEC (Figure 4C).
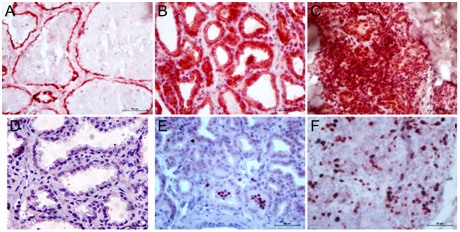
Figure 4 Immunostaining of MUC1 and Ki67 in goat mammary tissue. Expression of MUC1 in lactating tissue was present in cytoplasmic and apical boarder of alveolar cells (A). In mastitis, MUC1 expression was not remained confined to apical boarder but diffused all over the cytoplasm of alveolar cells (B). In cancer, MUC1 expression was high, aberrant (that is nuclear rather than cytoplasmic) and not limited to MEC but stromal cells as well (C). Sometimes minimal expression of MUC1 was observed and was associated with poorly differentiated MEC (not shown). .
Protein expression of cell proliferation marker, Ki67 did not differ between lactating and mastitic tissues (Figure 4A & 4B). However, its expression was significantly up-regulated in mammary carcinomas (Figure 4C). Mean percentage of Ki67-stained area (expressed in terms of integrated pixel density of Ki67-positivity) was high (14.5+2.2%).
Real time quantitative PCR
Expression of MUC1 was significantly upregulated (p=0.005) in cancer than that of mastitic tissue (Figure 5A). The mRNA expression of MUC1, FNDC3B (cancer cell markers), CCNB1, PCNA (cell proliferation markers) and MSI1 (mammary stem/progenitor cell marker) in lactating and cancer was examined using real-time reverse transcription quantitative PCR (RT-qPCR). FNDC3B, MUC1, CCNB1, MSI1 and PCNA was up-regulated 100 fold, 5-fold, 5-fold, 2-fold and 1.7-fold, respectively in cancer tissue than the lactating tissue (Figure 5B).
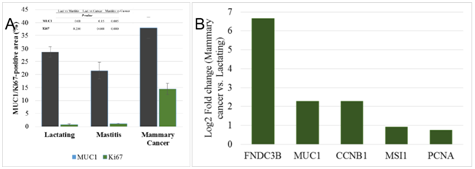
Figure 5 Expression analysis of cancer cell markers and cell proliferation markers in goat mammary glands. Immunohistochemical localization of MUC1 and Ki67 in lactating, mastitic and mammary cancers (A). Comparisons of MUC1 and Ki67 expression among three groups of animals with p-value is given. Fold change (log 2) of transcripts of cancer cell markers (FNDC3B and MUC1), cell proliferation markers (CCNB1 and PCNA) and mammary stem cell marker, musashi (MSI1) showed up-regulation in mammary cancer (B).
In this study, we characterized goat mammary cancers in terms of collagen fiber density and expression of molecular markers of cancer. We collected samples of mammary parenchymal tissue from abattoir and categorized them into various groups namely, lactating, mastitic and mammary cancers.
Picrosirius red staining is a simple, sensitive and specific method for collagen staining. It is particularly useful to reveal the molecular order and organization of collagen fiber orientation and anisotropy in connective tissues. In white-field microscopy, collagen fiber appears red and nuclei appear grey/brown. Collagen surrounding normal epithelial or stromal cells was loose, curly and smooth which turns to rigid, thick and more linear upon tumor development.12 Collagen density and organization are potentially key determinants of tumor cell behavior and they provide microenvironment to cancer stem cells for growth and spread. Role of collagen in recruitment of tumor associated macrophage during metastasis is already known.19
Immunolocalization of MUC1 and Ki67 was investigated in lactating, mastitic, and mammary cancer tissue in goats. MUC1 is a transmembrane mucin protein that is normally expressed by mammary epithelial cells during active secretion. It creates a wall between apical side of mammary epithelial cells and alveolar lumen thus preventing pathogen entry from outside to inside the cell. MUC1 often overexpressed in metastatic cancer and hence used as diagnostic marker of metastatic progression. Its extracellular domain serves as ligand for stromal and endothelial cells adhesion, while cytoplasmic domain is involved in several interactions and cell signaling that leads to migration and invasion. Overexpression of MUC1 has been demonstrated in various types of cancers including mammary carcinomas and hence considered as oncoproteins.20,21 Ductal and lobular carcinomas of mammary glands express sialylated MUC18 which is different from the MUC1 expressed in normal mammary gland and probably that could be reason for aberrant localization of the protein in cancer cells. Aberrant localized MUC1 therefore related with worst prognosis of the disease.10
A high Ki-67 expression is associated with aggressive carcinomas. High Ki67 expression is linked to unfavorable breast cancer outcomes in numerous breast cancer studies. In one such study, high Ki67 expression was associated with high grade cancer and poor prognosis.22 A negative correlation was found (r2=-0.21) between MUC1 and Ki67 expression in goat mammary carcinoma. Our result of negative correlation of MUC1 and Ki67 expression is not in agreement with the results published where no correlation of MUC1 and Ki67 is reported.11 This difference might be due to either species difference (goat vs. human mammary carcinoma), types of cancer or due to methods of analyzing the data.
Real time qPCR gene expression analysis has indication increased expression of cancer cell markers, cell cycle markers and proliferation markers. FNDC3B is mammary stem cell and cancer cell marker reported by many investigators.23–28 Our earlier report of increased expression of FNDC3B in buffalo mammary cancer24 was consistent with the present finding of goat mammary cancer. Cyclin B1 (CCNB1) is a cell cycle protein in G2/M transition. CCNB1 is also a prognostic marker of estrogen receptor positive breast cancer.29 Expression of all these markers in mammary cancer indicates core gene network.
In conclusion, mucin 1 was aberrantly expressed in goat mammary cancer. Consistent with the literature, aberrant and high expression of mucin 1 was associated with goat mammar cancer. Gene expression analysis of cell and proliferation markers was increased significantly in mammary cancer. Density of collagen fiber was high in cancerous tissue. This study is the first of its kind showing the detail cellular characterization of goat mammary cancer. Presence of high incidence of mammary cancer in goat is alarming and thus require detail investigation of its genesis. Further studies are needed to explain factors causing such high incidence of mammary cancer in ruminant.
Authors would like to acknowledge the Department of Biotechnology, New Delhi for the financial assistance (project reference number BT/AAQ/01/AB-I/TF-AP/2014).
The author declares no conflict of interest.

©2017 Choudhary, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.