Journal of
eISSN: 2475-5540


Review Article Volume 1 Issue 4
1Department of Hematology, Oncology and Molecular Medicine, Istituto Superiore di Sanit
2Institute of General Pathology, Universit
Correspondence: Lidia Villanova, Department of Hematology, Oncology and Molecular Medicine, Istituto Superiore di Sanit, Tel +39-0649902776
Received: July 28, 2016 | Published: August 22, 2016
Citation: Villanova L, Fiori ME. “De-coding” stemness pathways in cancer. J Stem Cell Res Ther. 2016;1(4):135-145. DOI: 10.15406/jsrt.2016.01.00024
The discovery of noncoding RNAs (ncRNAs) solved the apparent contradiction between the large extent of the human transcriptome and the relatively small number of protein-coding genes. MicroRNAs (miRNAs) and long noncoding RNAs (lncRNAs) account for the majority of noncoding transcripts with regulatory functions and proved to be important players in cancer onset and progression. Here we highlighted the oncogenic and tumor suppressive roles of ncRNAs, particularly focusing on how they affect cancer stem cells (CSCs)-sustaining signaling pathways. JAK-STAT, TGF-β, WNT and Hedgehog pathways control some major mechanisms fueling the cancer stem cell state, ranging from epithelial-to-mesenchymal transition (EMT) to the crosstalk with the tumor stroma. A further understanding of the mechanisms underlying ncRNAs function in CSCs will lay the foundation for developing therapeutic strategies more effective in selectively targeting the CSC population.
Keywords: microRNA, long noncoding RNA, cancer stem cells, EMT
ncRNAs, noncoding rnas; miRs, micrornas; lncRNAs, long noncoding rnas; RISC, rna-induced-silencing-complex; mRNAs, messenger rnas; CLL, chronic lymphocytic leukemia; RREB1, ras-responsive element-binding protein 1; HK2, hexokinase 2; AldoA, aldolase a; GPD2, glycerol-3-phosphate dehydrogenase-2; Xist, x inactive specific transcript; XCI, x chromosome inactivation; PRC2, polycomb repressive complex 2; PCA3, prostate cancer associated 3; AR, androgen receptor; FDA, food and drug administration; HOTAIR, hox transcript antisense rna; LSD1, lysine-specific histone demethylase 1; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; NEAT2, nuclear-enriched abundant transcript 2; HULC, highly up-regulated in liver cancer; SPHK1, sphingosine kinase 1; SP1, sphingosine-1-phosphate; MVIH, lncrna associated with microvascular invasion in hcc; PGK1, phosphoglyceratekinase 1; TERRA, telomeric repeat-containing rna; RPA, replication protein a; POT1, protection of telomeres 1; ALT, alternative lengthening of telomeres; ASOs, antisense oligonucleotides; ceRNAs, competing endogenous rnas; MREs, microrna recognition elements; CSCs, cancer stem cells; EMT, epithelial-to-mesenchymal transition; MET, mesenchymal-epithelial transition; lncRNA-ATB, lncrna-activated by tgf-β; HCC, hepatocellular carcinoma; GFP, green fluorescent protein; CTCs, circulating tumor cells; Shh, sonich hedgehog; Ptch, patched; Smo, smoothened; CCL21, c-c motif chemokine 21; CCR7, c-c chemokine receptor type 7; BCAR4, breast cancer anti-estrogen resistance 4; SNIP1, smad nuclear-interacting protein 1; PNUTS, serine/threonine-protein phosphatase 1 regulatory subunit 10; PP1, serine/threonine-protein phosphatase 1; CTD, carboxyterminal domain of rna pol ii; LNA, locked nucleic acid; LSCs, leukemic stem cells; Fz, frizzled; LRP5/6, low-density lipoprotein receptor-related protein 5/6; Dsh, dishevelled; TCF/LEF, t-cell factor/lymphoid enhancer factor; ESCs, embryonic stem cells; CML, chronic myeloid leukemia; EZH2, enhancer of zeste homolog 2; APC, adenomatous polyposis coli protein; CBP, creb-binding protein; CK1, casein kinase 1; DKK1, dickkopf-related protein 1; GAS1, growth arrest-specific 1; GSK3β, glycogen synthase kinase-3 beta; IL-11, interleukin-11; JAK, janus kinase; STAT, signal transducer and activator of transcription; TGF-β, transforming growth factor-β; TGF-βR, transforming growth factor-β receptor; ZEB1/2, zinc finger e-box-binding homeobox 1/2
In the last 15years the complete sequencing of Human Genome and its functional characterization revealed that only 20-25,000 protein coding genes are interspersed in about 2.85 billion nucleotides.1 In apparent contrast with these findings, the recent ENCODE project unveiled that about 76% of human genome is actively transcribed.2
It is nowadays well known that a plenty of noncoding RNAs are functionally relevant in both physiological and pathological conditions. Far from “one gene, one enzyme” paradigm, stated in 1945 by George Beadle, we now know that ncRNAs regulate gene expression at different checkpoints, enormously expanding the complexity of cell biology and the number of cellular adaptations to surrounding stimuli.
Here we will discuss advances in understanding the role of two major classes of ncRNAs: microRNAs (miRNAs, miRs) and long noncoding RNAs (lncRNAs), with particular focus on cancer and cancer stem cells.
microRNAs
MicroRNAs are small, non-coding RNAs of 18-25 nucleotides in length that are phylogenetically conserved. The first report on microRNAs dates back to 1993 when two papers on Cell described the post-transcriptional regulation of the lin-14 gene exerted by the lin-4 gene during larval development in Caenorhabditis elegans.3,4 After seven years, the discovery of let-7, another heterochronic small RNA involved in the developmental timing of C. elegans, broadened the interest of scientific community on these tiny molecules. Let-7 resulted extremely conserved in humans, flyes and nematodes and this finding paved the road for the discovery of about 2588 mature microRNAs in humans.5 From then on thousands of papers every year elucidated the role of miRNAs, described their biosynthesis and mechanisms of action in gene expression regulation. MiRs are generally transcribed by RNA Pol II as long primary transcripts (pri-miRs), that share common features with protein-coding transcripts. They are 5’ capped, polyadenylated and possibly contain introns that undergo splicing. A complex succession of processing events and intracellular shuffling leads to the final short molecule able to guide the RISC complex (RNA-Induced-Silencing-Complex) on target messenger RNAs (mRNAs) bearing imperfect complementary sequences. Upon miR binding, mRNA translation results impaired and/or mRNA degradation occurs.6–8
miRNAs as cancer drivers
The discovery in 2002 that a poly-cistronic cluster encoding for two microRNAs, miR-15 and miR-16, is often deleted in Chronic Lymphocytic Leukemia (CLL) suggested for the first time that deregulation of ncRNAs could contribute to human cancer onset.9 The oncosuppressive role of miR-15 and miR-16 was further demonstrated by the identification of one of their target genes, the oncogene BCL2.10 We now know that miRs behave as cancer drivers, being integrated in most oncogenic pathways. Accumulating evidence indicates that miRs govern all hallmarks of cancer.11 Since the extensive and deep description of the findings about microRNAs in cancer is not the main scope of this review, we will limit to some significant examples of miR deregulation in cancer, providing an overview of the multilayer integration of miRs in cancer-associated pathways.
The initial clues about possible roles of miRs as cancer determinants came from the analysis of differential expression in tumors compared to normal controls. miR-21 is the most over-expressed microRNA in different cancer types. It was firstly identified as aberrantly expressed in glioblastoma multiforme.12,13 In a large-scale study profiling miR expression in 363 specimen representing six different solid tumors versus 177 normal controls, miR-21 resulted the only miR up-regulated in all types of cancer analyzed (breast, colon, lung, pancreas, prostate and stomach).14 Other studies demonstrated high miR-21 expression in virtually all solid tumors and leukemic cancers.15–18 Additional reports demonstrated the power of miR expression profiles in stratifying cancer subsets and paved the road for the identification of cancer-specific miR-signatures, useful for both diagnostic and prognostic applications.19,20
Besides chromosomal amplifications or deletions, miR expression can be driven by tightly controlled steps, similarly to protein-coding genes. Transcription of miR-coding genes can be influenced by oncogenes or oncosuppressors. The first potentially oncogenic miR described, the miR-17-92 polycistron (also named oncomiR-1), is under the transcriptional control of the oncogene Myc.21–23 Mechanisms of miR-21 up-regulation at transcriptional level were also characterized. MiR-21 promoter was shown to be regulated by activation protein 1 (AP-1), composed of Fos and Jun oncogenes, whose activity is often potentiated upon oncogenic transformation.24 Furthermore, the augmented miR-21 levels in ovarian cell lines upon 5-aza-2'-deoxycytidine demethylating treatment, suggested that the expression of this microRNA is also under epigenetic control.17
Transcriptional regulation by key factors has been observed also for microRNAs endowed with oncosuppressive features. The oncosuppressor TP53 was shown to activate the transcription of a conserved microRNA family, the miR-34a-c, which in turn regulates the expression of target genes leading to cell cycle arrest and promoting apoptosis, thus contributing to TP53 activity.25 A number of reports have enriched the microRNAs/TP53 network, showing several microRNAs that directly target TP5326–29 and/or mediate TP53-dependent responses, as apoptosis induction.30
Suzuki and coworkers31 demonstrated that microRNAs synthesis is controlled not only at the transcriptional level. They showed that TP53 modulates microRNA processing by interacting with Drosha protein complex, namely with the RNA helicase p68 (DDX5). TP53 binding facilitates the maturation of miRs with growth-suppressive role, as miR-143/145 and miR-16-1, thus empowering the anti-tumor impact of TP53 network.31
The transcription of the oncosuppresive miR-143-145 cluster is repressed by KRAS, whose constitutive activation drives many cancer types. Kent and colleagues reported that ablation of miR-143 and miR-145 was necessary for tumorigenesis dependent on active KRAS pathway and unveiled that RREB1 is the KRAS effector that binds to and inhibits miR-143/145 promoter. Furthermore, they discovered a positive regulatory loop with KRAS and RREB1 being target of miR-143 and miR-145 in pancreatic cancer cells.32
An impressive number of scientific reports in the last decade described the role of hundreds of microRNAs in virtually all cancer types, unraveling the complex network of interactions that lead to fine tuning gene expression in a tissue-specific manner. Many targets were identified for every miR analyzed, in the attempt to explain the mechanisms of action of these tiny cancer determinants. MiRs proved to have a pivotal role also in regulating features recently described as novel hallmarks of cancer, such as communication of cancer cells with tumor stroma and metabolic reprogramming of cancer cells. Cancer cells are endowed with the ability to switch from oxidative phosphorylation to aerobic glycolysis and vice versa, adapting to variations in the microenvironment to survive hypoxia and promote glucose uptake to sustain active proliferation.33 A number of metabolic enzymes were found controlled by miRs, as glucose transporters (GLUT) by miR195-5p/miR-93, hexokinase 2 (HK2) by miR-155/miR-143, aldolase A (AldoA) by miR-15a/16-1 and glycerol-3-phosphate dehydrogenase-2 (GPD2) by miR-1 and miR-206.34–39
LncRNAs: New players on the RNA stage
Long noncoding RNAs are transcripts >200bp in lenght, commonly transcribed by RNA polymerase II. Their genomic context may greatly vary: some lncRNAs are transcribed from independent promoters or enhancers, some others from introns or antisense to other genes, or from pseudogenes.40 Although most lncRNAs undergo the same post-transcriptional modifications as messenger RNAs (mRNAs), including 5’ capping, 3’ polyadenylation and splicing, these transcripts often have no appreciable peptide product. Nevertheless, compelling experimental evidence, such as a tight regulation through well-established transcription factors, chromatin signatures and in some cases a tissue-specific expression pattern, argued for a biological role of lncRNAs. These observations pointed out lncRNAs as a novel class of transcripts with a strongly defined identity, thus ruling out that they could be a byproduct of a leaky transcriptional system in mammalian cells.
LncRNAs fold into secondary structures, exerting their functions through the interaction with other cellular macromolecules (chromatin, proteins, RNAs).41 At chromatin, lncRNAs can regulate expression of neighboring genes (in cis) or genes located on distant chromosomes (in trans) through multiple mechanisms: recruitment of chromatin modifiers on specific loci, establishment of genomic proximity between enhancer and promoter by chromosomal looping, transcription factor trapping, and others. In addition, lncRNA interaction with multiple proteins can facilitate or impair the assembly of protein complexes. Finally, lncRNAs are able to modulate mRNA processing at different levels (splicing, stability, translation) or act as decoys for miRNAs, buffering their inhibitory effect on the target mRNA.
One of the first functionally annotated lncRNAs was Xist (X inactive specific transcript),42 which mediates gene dosage compensation in female mammals. In fact, the difference in X-linked gene dosage between XX females and XY males is compensated by a mechanism known as X chromosome inactivation (XCI), in which one of the two X chromosomes in females is transcriptionally silenced, such that only one X remains active and is expressed in each female cell. Xist-mediated coating of the X chromosome to be inactivated serves as a scaffold for the recruitment of chromatin factors such as Polycomb repressive complex 2 (PRC2), which spread a heterochromatin state throughout the whole chromosome, resulting in its transcriptional repression.43,44 More recently, upon the advent of the unbiased high-throughput technology of RNA-sequencing, tens of thousands human lncRNAs were identified and these new versatile molecules gained the attention of the scientific community in the noncoding RNA field.
Functional role of lncRNAs in cancer
LncRNAs proved to play a major role in multiple cancers, where their expression levels are often disregulated. Some lncRNAs were found to be associated with common cancer-related genomic alterations, such as the amplification of the 8q24 locus, which occurs in many human malignancies and results in the amplification of the MYC proto-oncogene. For example, PVT1 lncRNA is adjacent to MYC locus and gain of PVT1 expression was shown to be required for high MYC protein levels in 8q24-amplified human cancer cells.45 Noteworthy, ablation of PVT1 from MYC-driven colon cancer cells impaired their tumorigenic potential, pointing out PVT1 as an attractive therapeutic target, especially as MYC protein has proved to be refractory to small-molecule inhibition.
The identification of thousands of lncRNAs differentially expressed in tumors relative to their normal tissue counterparts strongly suggested their potential as diagnostic biomarkers. The lncRNA PCA3 (prostate cancer associated 3) is currently used as an established cancer diagnostic biomarker. Originally discovered in 1999 upon analysis of the mRNA expression patterns of prostatic tumors versus adjacent non-neoplastic tissue of the same patients, PCA3 was described as a highly prostate-specific noncoding transcript.46 Unfortunately the biological function of PCA3 has remained elusive, besides few reports showing its role in androgen receptor (AR) signaling47 and in the regulation of a novel tumor suppressor in prostate cancer cells.48 Nevertheless, PCA3 was approved by the Food and Drug Administration (FDA) as a biomarker for prostate cancer diagnosis and its detection in the patients’ urine currently represents a useful noninvasive test.49,50
In addition, some lncRNAs are able to provide prognostic and predictive information. HOTAIR (HOX transcript antisense RNA) aberrant expression was shown to correlate with cancer progression in several tumor types and to predict differential sensitivity of ovarian cancer patients to two distinct chemoterapy regimen.51 The functional role of HOTAIR in platinum resistance of ovarian cancer has been further explored in a recent report. In response to cisplatin-induced DNA damage in ovarian cancer cell lines, HOTAIR is transcriptionally upregulated by NF-kB and mediates DNA damage response and cellular senescence, ultimately leading to chemoresistance.52
Here we will provide an overview of the biological functions of the major oncogenic and tumor suppressive lncRNAs in different cancer phenotypes or "hallmarks of cancer".11 Among the tumor-promoting lncRNAs, HOTAIR is the most well-characterized in multiple cancer types, where it exerts its function through different molecular mechanisms and impacts on distinct cancer phenotypes, ranging from increased proliferation to acquisition of cell motility and invasive potential. In prostate cancer, HOTAIR is a positive regulator of the proliferation circuit orchestrated by the AR signaling and its overexpression can drive androgen-independent prostate cancer growth and invasion. In fact, HOTAIR binding to the AR protein impairs its interaction with the E3 ubiquitin ligase MDM2, thereby preventing AR ubiquitination and degradation.53 In breast cancer, HOTAIR functions at chromatin where interacts with multiple targets, leading to activation of distinct tumor-promoting pathways. In a recent study, HOTAIR upregulation promoted ligand-independent chromatin occupancy of the estrogen receptor (ER) and potentiated its downstream gene regulation, ultimately contributing to tamoxifen resistance.54 Another report demonstrated that HOTAIR interacts with the histone-demethylase LSD1 at promoters of c-Myc target genes, thus mediating the activation of c-Myc transcriptional program.55 Moreover, HOTAIR was described to serve as a scaffold for the assembly of the chromatin-remodeling complex PRC2 (Polycomb repressive complex 2) at specific target genes. Interestingly, enforced expression of HOTAIR in epithelial cancer cells induced a genome-wide re-localization of PRC2, resulting in a switch in the chromatin landscape and gene expression pattern towards a more mesenchymal cellular phenotype. This phenotypic switch ultimately led to increased cancer invasiveness and metastasis.56
As the primary tumor grows, new vessels start sprouting from the existing vasculature in order to provide tumor cells with nutrients and oxygen supply, a process called angiogenesis. More than one oncogenic lncRNA was found to be involved in facilitating angiogenesis. Expression of MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), also referred to as NEAT2 (nuclear-enriched abundant transcript 2), was found upregulated in human neuroblastoma cells under hypoxic conditions and turned out to be necessary for FGF2-mediated induction of vasculature formation.57 Highly up-regulated in liver cancer (HULC) lncRNA promotes angiogenesis in hepatocellular carcinoma through a different molecular mechanism. HULC functions as a microRNA sponge, sequestering miR-107 away from its target, E2F1 mRNA, thus promoting E2F1-mediated transcriptional activation of SPHK1 (sphingosine kinase 1) and consequent activation of the SPHK1-SP1 (sphingosine-1-phosphate) axis, which enhanced angiogenesis in vitro and in tumor xenografts.58 MVIH (lncRNA associated with microvascular invasion in HCC) was also reported to induce angiogenesis in hepatocellular carcinoma, by directly associating with PGK1 (phosphoglyceratekinase 1) protein and reducing its extracellular levels. Consistently, patients with a higher MVIH expression level in primary HCC tissues showed a substantial down-regulation of serum PGK1.59 Overall, these findings shed light on the lncRNA-mediated regulation of tumor-driven angiogenesis, which involves multiple interactors and pathways.
Replicative senescence owing to the progressive shortening of telomeres, specialized DNA-protein complexes protecting the ends of chromosomes, represents a barrier that tumor cells need to overcome in order to gain unlimited proliferative potential. The most common strategy that cancer cells adopt to circumvent this hurdle is overexpression of telomerase, a specialized DNA polymerase responsible for the extension of telomeric DNA, almost absent in non-immortalized cells of adult tissues. Telomeric DNA, which consists of multiple tandem hexanucleotide repeats ending in a 3' single-strand overhang, is associated with the shelterin protein complex, which protects the ends of chromosomes from being recognized as sites of DNA damage. Telomeres were considered to be transcriptionally silent, until telomeric repeat-containing RNA (TERRA) molecules were discovered.60 These lncRNAs are transcribed from several subtelomeric loci and localize to telomeres. Changes in TERRA levels at telomeres during cell cycle progression orchestrate the sequential binding of replication protein A (RPA) and the shelterin component protection of telomeres 1 (POT1) to the 3' single-stranded DNA (ssDNA) overhang, thus coordinating telomere replication and "capping" respectively.61 Consistently, TERRA foci at telomeres decline towards late S phase facilitating transient RPA association with telomeric ssDNA, to allow DNA replication. When TERRA levels rise again after S phase, RPA is displaced by POT1 binding and telomeric ssDNA may remain capped until the arrival of the replication forks in the next S phase, ensuring the orderly switch of telomere replication and capping processes, necessary to preserve genomic integrity.
Importantly, a subset of human cancers relies on a telomerase-independent mechanism to overcome replicative senescence, which is called alternative lengthening of telomeres (ALT) pathway. ALT relies on homologous recombination to elongate telomeres and RPA is a key mediator of this process. Interestingly, in cancer cells relying on ALT the cell-cycle regulation of TERRA is compromised, allowing for persistent association of RPA with telomeres after DNA replication and consequent triggering of recombination. Impairment of the ALT pathway induced apoptosis in ALT-positive cancer cells, suggesting that specific inhibitors may represent an attractive therapeutic strategy.62
An additional feature of cancer cells is the ability to escape the anti-proliferative programs orchestrated by tumor suppressor genes. TP53 is a crucial player in most of these regulatory networks. In response to DNA damage, replicative stress, persistent oncogenic insults and other sources of cellular stress, p53 induces cell cycle arrest to allow for repair of the damage. Alternatively, if the damage cannot be fixed, sustained p53 activation triggers either apoptosis or senescence. LincRNA-p21 was reported to play a tumor suppressive role in mediating p53-induced cell cycle arrest or apoptosis upon DNA damage. One study elucidated lincRNA-p21-driven cis-regulation of its neighboring gene, p21.63 Mechanistically, lincRNA-p21 recruits the coactivator hnRNP-K at the promoter of p21, thus activating its transcription, and indirectly promotes the expression of a set of DNA damage-inducible PRC2-target genes. Mouse embryonic fibroblasts (MEFs) transfected with antisense oligonucleotides (ASOs) specifically targeting lincRNA-p21 showed a compromised G1/S checkpoint and a faster growth rate, a phenotype consistent with p21 loss of function. In another study, lincRNA-p21 and hnRNP-K act in concert to repress genes in the p53 transcriptional response, thereby ensuring proper apoptotic induction upon irreversible DNA damage.64
Oncogene-induced senescence consists of an irreversible proliferative arrest triggered by oncogenic insults in pre-cancerous cells, as a defense mechanism that constraints the progression of benign lesions. The senescence-associated transcriptional program involves the stable repression of proliferative genes regulated by E2F transcription factors and the re-activation of senescence-enforcing genes such as CDKN1A (alias p21) and CDKN2A (alias p16) that are repressed by polycomb repressor complexes (PRC). In proliferating cells, PANDA lncRNA recruits PRCs to inhibit the transcription of senescence-promoting genes and the modulation of its expression levels are responsible for cellular entry or exit from the senescence program.65
Pseudogenes have recently emerged as a novel class of lncRNAs. Pseudogenes represent DNA loci structurally similar to known genes but unable to encode functional proteins, owing to the presence of premature stop codons, frameshift mutations, insertions, or deletions. In fact, pseudogenes are products of tandem gene duplications or retrotransposition events that occurred during genome evolution, both of which created extra gene copies that are no longer under selective pressure. For this reason they have been considered for a long time as useless evolutionary relics. Surprisingly, recent evidence revealed that many pseudogenes are transcribed and their sequences are often evolutionary conserved, suggesting that they may exert noncoding functions. Interestingly, Poliseno and colleagues proposed a new role for pseudogene transcripts, showing that they can behave as competing endogenous RNAs (ceRNAs). In fact, because of their sequence similarity, pseudogenes share multiple microRNA recognition elements (MREs) with their parental genes and can compete for the binding of common microRNA molecules. As a result, pseudogenes sustain the expression of their parental genes and hence can acquire oncogenic or oncosuppressive functions when deregulated. PTENP1 was the first pseudogene acting as a ceRNA to be identified in human cancer cells.66 Its 3’ UTR exerts a tumor suppressive function by acting as a sponge for PTEN-targeting miRNAs, hence increasing PTEN levels. Consistently, the PTENP1 locus is selectively lost in human cancer, providing a proof-of-principle of its tumor suppressive role. After the discovery of PTENP1, the list of oncogenic and oncosuppressive pseudogenes functioning as ceRNAs for their parental genes has been greatly expanded.
Cancer is not a unique pathology as every tumor type is characterized by specific driver and passenger mutations as well as epigenetic alterations. On top of this complexity, intra-tumor heterogeneity is fostered by coexistence within the same tumor of sub-clones featuring different genetic and epigenetic modifications.67 This heterogeneity represents a great challenge for current medicine: the identification of effective therapies is not straightforward as different clones display substantial functional diversity that accounts for recurrence and resistance to therapy. Moreover, cancer cells within virtually every clone can be further classified on the basis of non-heritable traits, giving rise to a hierarchical organization of the tumor. The bulk of a tumor is composed mainly by partially differentiated cells, endowed with a limited proliferative potential, while a sub-population of cancer cells, usually a small fraction of the whole, named Cancer Stem Cells (CSCs), displays features typical of normal stem cells, such as self-renewal, capacity to undergo asymmetric division and elevated resistance to apoptosis.68 Whether CSCs arise from normal stem cells that acquired cancer driver mutations or are consequence of de-differentiation of tumor cells is still on debate.68,69 Cancer cells stemness is increasingly viewed as a dynamic condition highly dependent on a combination of genetic, epigenetic and environmental conditions, rather than a cell-intrinsic state.70 The recent awareness of CSCs plasticity doesn’t change the principle that stemness determinants (whether transiently acquired or permanently expressed) are responsible for cancer recurrence, dissemination and therapy failure, candidating this tumor sub-population to be the best therapeutic target to pursue.
CSCs express mesenchymal traits, typical of cells that underwent epithelial-to-mesenchymal transition (EMT). The EMT program in non-pathological conditions has a pivotal role in development and tissue homeostasis. It is governed by several convergent signal transduction pathways that cooperate to integrate environmental stimuli. In the tumor context autocrine and paracrine factors secreted both by tumor cells and non-cancerous tumor-associated cells of the stroma (fibroblasts, endothelial cells, immune cells) contribute to activate and sustain the EMT program in CSCs. The main pathways controlling EMT will be described in this section that will provide an overview of stemness regulation triggered by ncRNAs.
ncRNAs in CSCs programs
During EMT, epithelial cells lose cell polarity and cell-cell adhesion and acquire motility, migratory capacity and invasive power, thus gaining the ability to colonize distant organs and initiate metastases.
NcRNAs are widely understood to be critical regulators of the metastatic process. An outstanding example is miR-10b which has proven to induce metastasis in breast cancer mouse models and is associated with several high grade malignancies. MiR-10b synthesis is induced by TWIST, a transcription factor promoting EMT, and it exerts its pro-metastatic function mainly through direct targeting of HOXD10.71 Weinberg and colleagues provided strong evidence that miR-10b is a promising candidate as therapeutic target, showing how delivery of a miR-10b antagonist in vivo was sufficient to repress breast cancer spread to metastatic sites,72 while leaving onset and development of primary lesions substantially unaffected.
Another miR that was shown to be involved in the EMT program is the p53-transcribed miR-34 that inhibits the translation of the EMT-promoting transcription factor SNAIL. Moreover, miR-34 transcription is inhibited by TGF-β induced SNAIL and ZEB factors that directly recognize E-Boxes in its promoter, giving rise to a negative feedback loop.73,74 Furthermore, Rocavec and colleagues demonstrated another regulatory loop involving miR-34 which is determinant for EMT induction in colorectal cancer. They showed that miR-34 expression is repressed by IL-6-induced STAT3, and that IL-6 receptor (IL-6R) is a direct target of miR-34.75
Two scientific reports in 2011 demonstrated that the oncosuppressor TP53 controls stemness plasticity by inducing mesenchymal to epithelial transition (MET) in mammary and hepatocellular carcinoma cells.76,77 This anti-metastatic activity of TP53 is exerted through miR-mediated regulation of ZEB transcription factors. In particular, TP53 directly promotes transcription of miR-200 family that then target ZEB1/2.78,79 ZEB factors are EMT effectors activated by TGF-one of the major signaling cascades involved in stemness control, and actively repress, among others, E-cadherin gene and miR-200.80–82
Strong evidence supporting the role of long ncRNAs in the EMT program came from the study published from Yuan and collaborators.83 The authors identified a novel lncRNA, named lncRNA-ATB (lncRNA-activated by TGF-β), whose expression was induced following TGF-beta treatment of hepatocellular carcinoma (HCC) cell lines. Their work is a remarkable example of how the same lncRNA can affect multiple steps of the invasion-metastasis cascade by modulating different cancer stem cell pathways. First of all, lncRNA-ATB sequesters miR-200 functioning as a ceRNA, thus relieving ZEB1 and ZEB2 mRNAs from miR-200 post-transcriptional inhibition. The resulting increased levels of ZEB1 and ZEB2 trigger the EMT program, ultimately leading to an enhanced invasion potential of HCC cells in vitro and in vivo. In fact, orthotopic injection of lncRNA-ATB-overexpressing clones resulted in increased intrahepatic, mesenteric, pulmonic and diaphragmatic metastases. However, the role of lncRNA-ATB in cancer metastasis goes beyond its impact on tumor invasion, which is only the early step of the invasion-metastasis cascade. To gain further insight into the later steps of this process, the authors investigated the function of lncRNA-ATB in the entrance of cancer cells into the circulation, by measuring the number of GFP-labeled circulating tumor cells (CTCs) following orthotopic injection of HCC clones overexpressing or depleted of lncRNA-ATB. Importantly, ectopic expression of lncRNA-ATB strongly increased the number of CTCs, and this effect was dependent on the competitive binding of miR-200.
Furthermore, lncRNA-ATB proved to be an important player also in the last step of the invasion-metastasis cascade, that is metastatic colonization in distant organs. The authors exploited two different experimental models (liver colonization upon intrasplenic injection and lung colonization following tail vein injection) to show the ability of lncRNA-ATB to promote metastatic colonization of HCC cells. Notably, this effect was not dependent on miR-200 inhibition or EMT, but involved an additional pathway crucial for the maintenance of the cancer stem cell state, the JAK/STAT signaling. According to the proposed model, lncRNA-ATB physically associates with the mRNA of IL-11, a known target of TGF-β pathway, thus increasing its stability. The resulting increase in IL-11 translation and secretion activates STAT3 signaling in an autocrine fashion. The activation of this autocrine loop is required for the boosting effect of lncRNA-ATB on the colonization potential of HCC cells. Overall, lncRNA-ATB activity is paradigmatic of how a ncRNA can modulate different cancer-related pathways in a synergistic fashion, resulting in an effective and multifaceted impact on tumor progression.
Another crucial pathway for the maintenance of the CSC state is Sonic hedgehog (Shh) signaling.84 The canonical pathway is triggered by the binding of secreted Shh ligand to the transmembrane receptor Patched (Ptch). In the absence of ligand, Ptch constitutively represses the activity of another transmembrane protein, Smoothened (Smo). Following Shh binding to Ptch, the repression of Smo is released, resulting in the transcription of target genes, mediated by the GLI family of transcription factors. Zhou and collaborators identified a novel lncRNA, named lncRNA-Hh, upregulated in breast cancer cells that had undergone EMT and acquired a CSC-like phenotype upon Twist ectopic expression. According to the proposed model, lncRNA-Hh is transcriptionally induced following Twist overexpression and triggers the activation of Hedgehog signaling, likely via the in silico predicted target GAS1 (Growth arrest-specific 1).85 GAS1 cooperates with Ptch for Shh ligand binding on the cell surface, thus acting as an enhancer of Shh signaling.86 Pathway activation was shown to increase the expression of GLI1 and the downstream targets SOX2 and OCT4, known master regulators of pluripotency and stemness. Accordingly, lncRNA-Hh knockdown in Twist-overexpressing breast cancer cells impaired tumorspheres-forming ability, self-renewal capacity in vitro, and tumorigenic potential in mouse xenografts.
Importantly, the EMT program and possibly the entrance of carcinoma cells into the CSC state are triggered by extrinsic signals arising in the tumor-associated stroma. The concept of tumor stroma has gradually evolved from the idea of a static environment to a dynamic interactor, actively promoting tumor growth and progression. The main constituents of the tumor microenvironment are the extracellular matrix, cells of hematopoietic origin of both the lymphoid and the myeloid lineage (such as lymphocytes, neutrophils and macrophages) and cells of mesenchymal origin (including myofibroblasts, adipocytes and endothelial cells). Each stromal cell type affects the tumor by secreting factors that induce the formation of cancer stem cells and help maintain the stemness state, often promoting cell migration and invasive ability. One example is CCL21-CCR7 paracrine signaling, which was shown to play a critical role in enhancing the metastatic ability of tumor cells. Indeed, high expression of the chemokine receptor CCR7 was detected in primary breast carcinomas and CCR7-mediated signaling triggered actin polymerization and pseudopodia formation in breast cancer cells, inducing chemotactic and invasive responses.87 In a recent report, the release of CCL21 chemokine from lymphatic endothelial cells acted in a paracrine fashion to mediate chemotactic migration of breast cancer cells undergoing EMT toward lymphatic vessels.88 Noteworthy, Xing and colleagues unveiled the role of lncRNA BCAR4 (breast cancer anti-estrogen resistance 4) in mediating chemokine-induced non-canonical Hedgehog signaling, promoting breast cancer metastasis.89 Indeed, in response to CCL21-CCR7 crosstalk, BCAR4 is recruited to the promoters of GLI2 target genes, where triggers transcriptional activation by interacting with two different nuclear proteins, via distinct regions of the transcript. On one side, BCAR4 binds to SNIP1, thus releasing the inhibition of the histone acetyltransferase p300 that results in the acetylation of specific histone marks required for gene activation; on the other side, BCAR4 interacts with PNUTS, whose binding to the acetylated histone tails induces the release of the inhibitory effect on PP1 activity. PP1-mediated dephosphorylation of the carboxyterminal domain (CTD) of RNA Pol II, bound to the promoters of GLI2 target genes, allows for the start of transcription. Finally, in vivo BCAR4 targeting through intravenous administration of a sequence-specific locked nucleic acid (LNA) resulted in reduced lung colonization of breast cancer cells. Overall, this elegant work provided strong evidence for the role of a lncRNA in the translation of the signals derived from the tumor stroma into the metastatic behavior of cancer cells.90
Wnt signaling pathway is an evolutionarily conserved pathway that regulates both stem cell maintenance and tumorigenesis. Wnt ligands are secreted glycoproteins that bind to the extracellular domain of Frizzled (Fz) receptor family and to co-receptors low-density-lipoprotein-related protein5/6 (LRP5/6). Upon ligand binding, the signal is transduced into the cytoplasm through the binding of Fz with the protein Dishevelled (Dsh) that leads to inactivation of β-catenin destruction complex. The adherens junction associated-protein β-catenin is then accumulated in the cytoplasm and translocates into the nucleus, where it activates the Wnt-driven transcription program in concert with T-cell factor/lymphoid enhancer factor (TCF/LEF) and other co-factors. Among Wnt target genes, a miRNA cluster, the miR-371-373 cluster, was shown to be specifically expressed in embryonic stem cells (ESCs) and frequently upregulated in several human cancers. Zhou and colleagues demonstrated that these Wnt-activated miRs in turn foster Wnt signaling by targeting the secreted negative regulator of Wnt Dickkopf (DKK1).91
In another study a lncRNA, lncTCF7, was found to play a major role in triggering self-renewal of CSCs in hepatocellular carcinoma (HCC).92 In this report, cancer stem cells were isolated from HCC cell lines and primary samples according to the expression of the surface markers CD13 and CD133. LncTCF7 depletion in the CD13+CD133+ cell fraction significantly reduced the expression of pluripotent transcription factors Sox2, Nanog, and Oct4 and reduced the formation of tumorspheres and tumor initiating capacity in mouse xenografts. Concomitantly, lncTCF7 silencing dramatically down regulated expression of the transcription factor TCF7 and major Wnt targets. The authors demonstrated an elegant mechanistic model where lncTCF7 directly binds to the chromatin remodeling SWI/SNF complex via a stable stem-loop structure in the 3’ end fragment. This interaction increases the occupancy of the SWI/SNF complex at TCF7 promoter, thereby leading to enhanced TCF7 transcription and, ultimately, to priming of the self-renewal of liver CSCs and tumor initiation. Interestingly, knockdown of lncTCF7 resulted in decreased expression of some major Wnt ligands, including Wnt7a, Wnt4, and Wnt2b, suggesting a role for this lncRNA in the maintenance of the CSC state through cell-autonomous secretion of autocrine signals.
A very recent report of the same group underlined the role of another lncRNA, named lncRNA for β-catenin methylation (lnc-β-CATM), in the modulation of Wnt pathway.93 Lnc-β-CATM was identified as overexpressed in CD13+CD133+ liver CSCs isolated from HCC patients, compared to non-stem cell population and its accumulation in HCC patients reflected tumor grade classification. The authors demonstrate that lnc-β-CATM is required for liver CSCs self renewal. The proposed mechanism of action depends on the interaction of lnc-β-CATM with β-catenin and enhancer of zeste homolog 2 (EZH2), a key component of Polycomb repressive complex 2 (PRC2). EZH2 is a histone methyltransferase that mediates also methylation of non-histone proteins. According to the findings of this paper, lnc-β-CATM promotes β-catenin methylation by EZH2, suppressing its ubiquitination and consequent degradation, ultimately potentiating Wnt signaling.
CSCs have been identified as the subpopulation responsible for tumor relapse, owing to the ability to resist cytotoxic therapy and propensity to form metastases. One of the major challenges of current therapeutic efforts is to eradicate this subpopulation. In fact, the use of a selective targeted therapy hitting CSCs would attenuate their ability to survive conventional cytotoxic therapies and cause relapse and eventually impair or delay the metastatic spread of the tumor. Conventional chemotherapies could be then employed as a second-line approach, aiming at killing the transient-amplifying progenitors and the more differentiated cells constituting the bulk of the tumor.
Deregulation of the signaling pathways described in the Figure 1 leads to aberrant mechanisms sustaining CSC self-renewal by fostering EMT and autocrine signals that are crucial for engaging the further steps of tumor progression. In fact, once the EMT program and the CSC state are induced in carcinoma cells from paracrine signals coming from the tumor stroma, positive feedback loops originating from the activated Wnt and TGF-β signaling pathways give rise to autocrine signals that serve to maintain the stemness features in the absence of further paracrine stimuli. These self-sustaining mechanisms are crucial during invasion and metastasis, when CSCs maintain their mesenchymal traits in the absence of the signals arising from the primary tumor microenvironment. Hence the importance to hit molecular targets which impact both on intrinsic properties of CSCs, such as positive autocrine loops, self-renewal, asymmetric division ability and apoptosis resistance, and on extrinsic elements such as the signals arising from the tumor stroma.
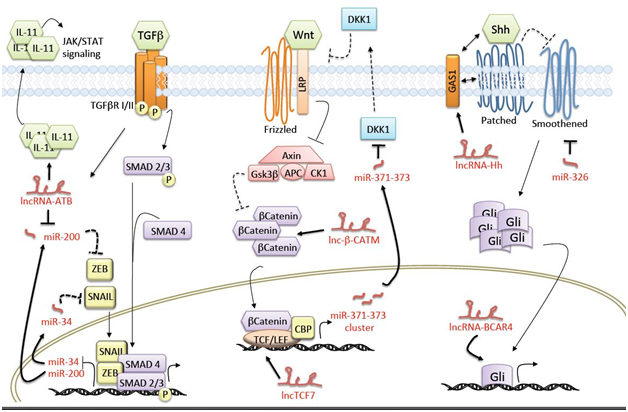
The figure illustrates the role of microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) in regulating key pathways involved in triggering and maintaining the epithelial-to-mesenchymal transition (EMT) program and the cancer stem cell (CSC) state.
Importantly, we are now aware that the majority of the transcriptome exerts a regulatory function on a relatively small number of protein coding genes. This complex regulatory network greatly expands the number of potential molecular targets to develop anti-CSCs therapies. To this purpose, it is crucial to identify the intersections between ncRNAs functions and CSCs-sustaining signaling pathways.
A wide range of cutting-edge technologies is currently available and allows a deep understanding of microRNAs interactome. More recent applications aim at identifying lncRNAs binding partners. Among others, RNA-protein (i.e. crosslinking immunoprecipitation, CLIP,94 RNA-DNA (i.e. capture hybridization analysis of RNA targets, CHART,95 chromatin isolation by RNA purification, ChIRP96 and RNA antisense purification, RAP97 or RNA–RNA (i.e. crosslinking analysis of synthetic hybrids, CLASH) interaction assays. Of these technologies, ChIRP-like methods are the most versatile, allowing the concurrent isolation of lncRNA protein interactors, genomic binding sites and RNA cofactors.98 Importantly, the in vivo purification strategy allows the analysis of physiologic interactions. Briefly, the ribonucleoprotein (RNP) complexes are cross-linked in vivo and solubilized by sonication. Sequence-specific biotinylated oligonucleotides are then hybridized to target the lncRNA of interest, which is efficiently pulled down with streptavidin magnetic beads together with the associated RNA molecules, DNA fragments and protein complexes. High-throughput sequencing and mass spectrometry coupled to ChIRP enable the simultaneous analysis of the genomic occupancy and of the RNA and protein interactomes of lncRNAs, providing a powerful tool for the study of lncRNA impact on several pathways at multiple layers of regulation. This approach is particularly applicable to the study of cancer stem cells biology, which relies on the crosstalk between different pathways and layers of regulation.
An understanding of the functional importance of lncRNAs in the context of the whole organism still relies on manipulating their expression by genetic modification, overexpression or knockdown strategies, and analyzing the resulting phenotypes. Although different strategies for analysis of lncRNA loss of function have been used, the mechanistic dissection of these putative regulators in vivo is still technically challenging.99 RNA interference (RNAi)-based techniques trigger transcript degradation in the cytoplasm, thus being useless for those lncRNAs that predominantly localize to the nucleus, such as those functioning at chromatin. On the contrary, antisense oligonucleotides (ASOs) can downregulate lncRNAs in the nucleus, by forming a DNA/RNA hybrid that triggers RNase H-dependent degradation. However, off-target effects cannot be ruled out. Also, it is not possible to generate stable transgenic lines or animals, which restricts analysis to experimental models where the oligonucleotides are supplied by injection. An alternative experimental approach is to genetically manipulate the lncRNA locus by deletion or inversion of either the promoter or the whole locus, or insertion of a transcription terminator. These manipulations can cause unintended changes in gene expression owing to multiple mechanisms: for instance, deletion of the lncRNA promoter could impair the expression of a neighbor gene sharing the same regulatory sequence. Further, other DNA regulatory elements can inadvertently be inserted or removed and the epigenetic environment can be perturbed. Unexpected gain of function effects can potentially arise from these genetic manipulations and strongly impact on the resulting phenotypes.
One interesting application of the lately developed genome editing technology CRISPR exploits short guide RNAs (sgRNAs) to target repressor or activator domains to any genomic location, including lncRNAs loci, to affect transcriptional initiation. This system has the advantages to not making permanent alterations to the genomic DNA and to be customizable. In fact, fusing the short guide RNA to any modular enzymatic component enables recruitment of a variety of desired perturbations to a locus of interest.100 In addition, this approach enables efficient targeting of multiple genomic sequences in parallel.101 However, not all the aforementioned strategies can easily separate roles performed by an RNA transcript from those performed by its corresponding DNA locus or potentially encoded peptide.
Shechner and colleagues recently described a novel approach to study lncRNAs function, called CRISPR-Display.102 Interestingly, the type II CRISPR-Cas9 system is engineered to deliver long structured RNAs, instead of enzymatic components, to a locus of interest. This goal is achieved by the incorporation of the desired long RNAs into the sgRNA, which targets a nuclease-deficient Cas9 mutant to a specific locus. Interestingly, this approach is multiplexable, enabling targeting of a lncRNA of interest to multiple genomic sites. The potential applications of this tool range from basic research, for the study of natural lncRNAs functions allowing uncoupling of RNA activity from its transcription, to synthetic biology and translational research, enabling the construction of novel lncRNA-based devices.
Overall, we can now count on a powerful “toolkit” for the study of ncRNAs functions, which will pave the way to novel targeted therapies against CSCs. Noteworthy, miR-based therapies are already in clinics for Hepatitis C (miravisen, Santaris Pharma and RG-101, Regulus Therapeutics) while a Phase I clinical trial is currently ongoing for miR-34a mimic in haematological malignancies, liver cancer and solid tumors. The development of the CRISPR-Display technology has set the foundation for the generation of lncRNA-based therapeutics. We hope that many of the exciting discoveries regarding lncRNAs will be translated in effective therapies for patients in the next future.
The authors thank the Italian Foundation for Cancer Research (FIRC) for the fellowship award to L. Villanova.
The author declares no conflict of interest.

©2016 Villanova, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.