Journal of
eISSN: 2373-4426


Case Report Volume 5 Issue 2
1Nationwide Children's Hospital and the Ohio State University College of Medicine, USA
2Dartmouth-Hitchcock Medical Center and Geisel School of Medicine at Dartmouth College, USA
3Cincinnati Children's Hospital Medical Center and the University of Cincinnati College of Medicine, USA
Correspondence: Stephanie L. Santoro, MD, Nationwide Childrens Hospital and the Ohio State University College of Medicine, 700 Childrens Drive, Columbus, OH 43205, USA, Tel (614) 722-3535, Fax (614) 722-3546
Received: August 12, 2016 | Published: August 31, 2016
Citation: Santoro SL, Bao L, M. Saal H (2016) Terminal 14q Deletion and Duplication with Gastrointestinal and Pulmonary Disease. J Pediatr Neonatal Care 5(2): 00174. DOI: 10.15406/jpnc.2016.05.00174
Terminal deletions of 14q are rare but have typical clinical findings whereas distal duplications of 14q are less well characterized. The combination of terminal deletion and distal duplication of 14q has only been reported once before. Neither terminal deletions nor duplications are consistently reported to have gastrointestinal or pulmonary manifestations. We report a terminal deletion and a distal inverted duplication of 14q in a patient with multiple anomalies; duodenal malrotation, feeding intolerance, cholestasis, tracheo-laryngo-pharyngomalacia and small bilateral congenital cystic adenomatoid malformations with severe obstructive sleep apnea. This is the first report of gastrointestinal and pulmonary features in a patient with both terminal 14q deletion and distal 14q duplication.
Keywords: chromosome 14q, duplication, chromosome 14q, terminal deletion, Cytogenomic array, dysmorphic facial features
DEL, deletion; DUP, duplication; NAHR, non-allelic homologous recombination; HIDA, hydroxy iminodiacetic acid; GI, gastrointestinal; CPAM, congenital cystic adenomatoid malformation
With only twenty reported cases, features of the rare 14q terminal deletion syndrome include developmental disabilities, microcephaly, growth delay, hypotonia and varied dysmorphisms.1-4 Ophthalmologic, cardiac, neural, renal and genitourinary anomalies are associated.5,6 Rare features include limb anomalies, recurrent otitis media and seizure disorder.1,6 In contrast to 14q terminal deletion syndrome, duplication of chromosome 14q is more rare and not well-characterized. Pertinent features include dysmorphic facial features, growth delay, cardiovascular, neurologic, renal, genitourinary, limb, hearing, vision and immune system anomalies and development disability ranging from moderate to global impairment.7-11
Gastrointestinal (GI) and respiratory diseases are variable in 14q deletions and duplications. Atresias (anal, esophageal and tracheoesophageal), imperforate anus, midgut malrotation, intestinal dysmotility, pyloric stenosis and laryngomalacia have been identified but there is no consistent phenotype with 14q aberrations.4,8,12-16 Terminal deletion 14q and duplication 14q in a patient with neonatal hypotonia, psychomotor retardation, intellectual disabilities, short stature and facial dysmorphism has been reported.17 We describe a patient who has both terminal deletion and distal duplication of 14q with multiple anomalies involving the gastrointestinal and pulmonary systems.
The female patient was the product of a 34 week vaginal delivery to a healthy 19 year old primagravid woman and her unrelated partner with unremarkable prenatal history, no pregnancy complications and unknown etiology for preterm labor. APGAR scores were 5 and 9 at 1 and 5 minutes respectively; birth weight was 1580 g (3rd centile), length was 41.5 cm (5th centile) and head circumference was 30 cm (25th centile). At 5 days of age, genetics consultation was performed for dysmorphic facial features. The family history revealed a maternal half uncle and maternal cousin with cognitive impairment. No congenital anomalies, recurrent pregnancy loss or stillbirth were known but history from the paternal side was minimal. Physical exam showed a female infant with a prominent anterior fontanelle, clinodactyly and dysmorphic features including short palpebral fissures, epicanthal folds, a thin upper lip, a short upturned nose and micrognathia (Figure 1).
At initial hospitalization, medical issues were numerous and involved multiple organ systems. Persistent feeding intolerance required gastrostomy tube placement; Nissen fundoplication and Ladd’s procedure were performed for intestinal malrotation. Cholestasis from bile duct obstruction developed and paucity of bile ducts were seen on liver biopsy. Severe obstructive sleep apnea, periodic respirations and central apnea were noted on polysomnography. CT angiogram and microlaryngoscopy and bronchoscopy demonstrated significant glossoptosis, retrognathia, congenital cystic adenomatoid malformation (CPAM), tracheomalacia and severe pharyngo-laryngomalacia necessitating tracheostomy. Bilateral pelviectasis and small kidneys were detected on renal ultrasound. Cardiac anomalies included a bicommisural aortic valve, a small atrial septal defect and a patent foramen ovale. Brain MRI showed delayed sulcation, prominence of the subarachnoid spaces and delayed myelination for age.
Banded chromosome analysis was performed on peripheral blood according to standard cytogenetic procedure. Karyotyping revealed additional material on the distal portion of the long arm of chromosome 14 or 46, XX, add(14)(q31) (Figure 2). Parental cytogenetic studies were normal. Genomic DNA was extracted using Magna Pure Compact automated extraction kits (Roche, USA) following manufacturer’s instruction. Microarray analysis was performed using the Illumina HD Human OMNI-quad Bead Chip array following the manufacturer’s instruction (Illumina Inc., San Diego, CA, USA) and the microarray results were analyzed using the Illumina Genome Studio V2011.1 software and gene annotations were determined using NCBI build 36/hg18.
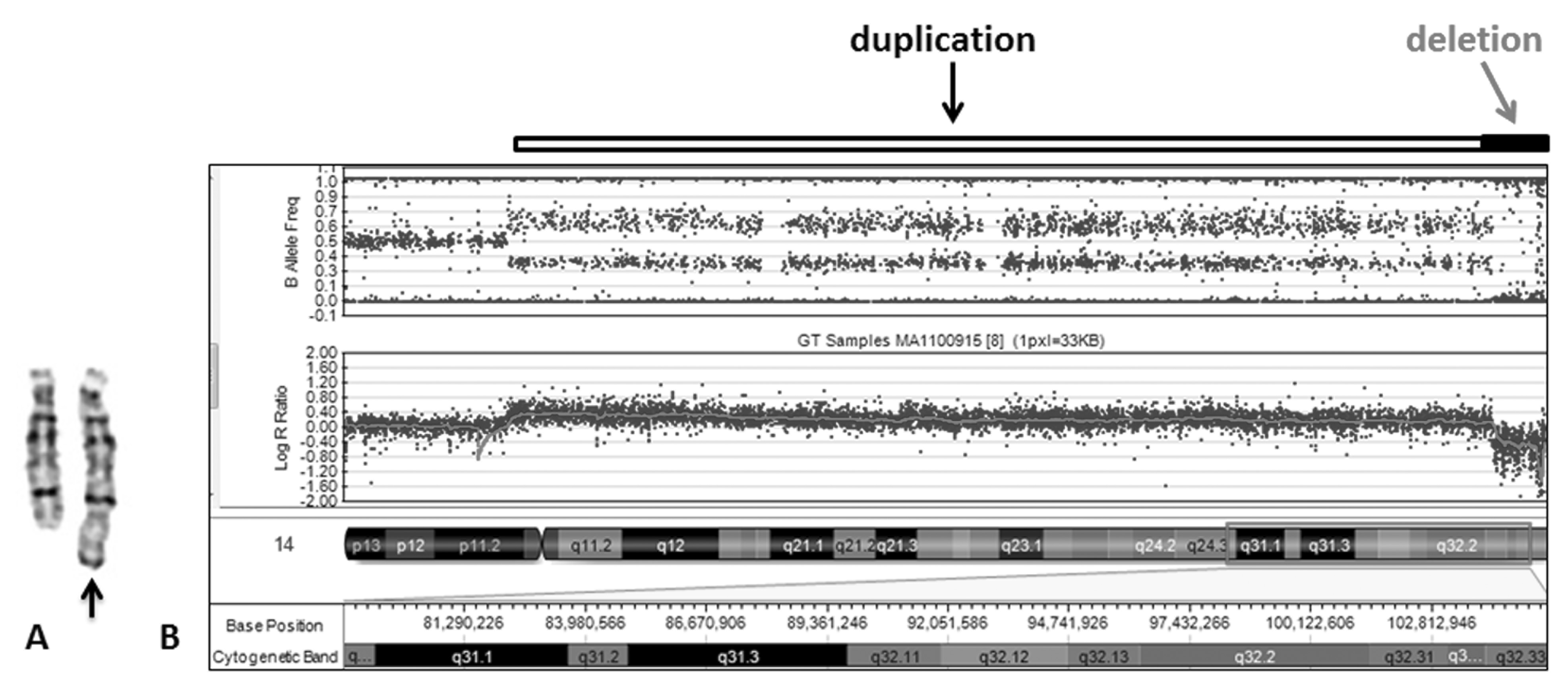
Figure 2 karyotyping and cytogenomic array study. (A) Partial karyotype showing a normal and abnormal chromosome 14 (arrowed). (B) Cytogenomic array analysis of chromosome 14 showing duplication of 14q31.1q32.33 and deletion of 14q32.33 as indicated by open and solid bars, respectively.
Microarray showed a large duplication spanning the bands 14q31.1 to 14q32.33 on the long arm of chromosome 14 with the linear positions of chr14:82,224,425-104,139,172 (21.9 Mb) and a small deletion at the band 14q32.33 with the linear positions of chr14:104,151,110-106,360,098 (2.2 Mb) (Figure 2). The duplicated region contains the disease genes including XRCC3, AMN, DYNCIH1, SPATA7, GALC, VRKI, TCLIA, TDPI, GALC, GOLGA5, TDP1, TTC8, SLC24A4, GLRX5, DICER1, SERPINA3, and ATXN3 and the deleted region has disease genes AKT1 and INF2. Thus, microarray uncovered the abnormal chromosome 14 observed in chromosome analysis which had a duplication and a submicroscopic deletion; the karyotype was revised to 46,XX,der(14)dup(14)(q32.33q31.1)del(14)(q32.33).
Following stabilization of medical issues, she returned for follow-up at 9 months of age without additional medical concerns and with developmental progress: she could hold her head up independently, sit with support, reach for objects and make noises. Additional dysmorphic features were noted including midface retrusion, broad forehead, hypertelorism, low-set ears, low nasal bridge with hypoplastic alae nasi, single palmar creases, brachydactyly, nystagmus and hypotonia (Figure 3). At 4 years of age, she had severe global delays, with more significant gross motor delays (Figure 4). She developed contractures of elbows and knees most likely related to increased appendicular tone. She did not eat and required gastrostomy tube feeds. Institutional review board at the Cincinnati Children’s Hospital Medical Center reviewed and approved the study.
Chromosome 14q abnormalities have been associated with various clinical features including inconsistent gastrointestinal and respiratory defects.7-19 Phenotypic variability can be attributed to differences in size of the 14q rearrangment, an additional chromosomal abnormality or the parental origin of the additional 14q segment.20-22 Although genomic imprinting effect has been reported in chromosome 14, such effect has not been reported in distal 14q.19,23 Molecular cytogenetic analysis showed both terminal deletion and distal duplication. Although one case with distal 14q inverted duplication and terminal deletion was reported previously, our case is the first to be characterized with high density microarray to delineate the abnormality at the molecular level (Table 1).17
Features |
A Terminal Deletion of 14q32.3 to qter* |
A Duplication of 14q31 to qter** |
Chen2 |
This case |
No of cases |
7 |
9 |
||
Male |
2 |
5 |
||
Female |
5 |
4 |
||
Mental retardation |
7/7 |
7/7 |
+ |
+ |
Hypotonia |
5/7 |
5/9 |
+ |
+ |
Growth retardation |
3/7 |
7/8 |
+ |
+ |
Microcephaly |
4/7 |
6/9 |
+ |
- |
Clinodactyly |
0/2 |
7/9 |
+ |
+ |
Hypertelorism |
1/4 |
7/9 |
+ |
+ |
Strabismus |
3/5 |
1/7 |
- |
- |
Blepharophimosis |
5/6 |
0/7 |
+ |
- |
Ptosis |
4/7 |
0/7 |
- |
- |
Downslanting palpebral fissures |
3/6 |
7/9 |
+ |
- |
Epicanthi |
5/7 |
2/9 |
+ |
+ |
Dysmorphic nose |
6/6 |
6/9 |
+ |
+ |
High arched palate |
5/6 |
3/9 |
- |
- |
Abnormal ears |
2/6 |
7/9 |
+ |
+ |
Micrognathia |
3/6 |
8/9 |
- |
+ |
Congenital heart defect |
2/6 |
6/9 |
+ |
+ |
Umbilical and diaphragmatic hernia |
0 |
1/9 |
- |
- |
Hiatal hernia |
0 |
1/9 |
- |
- |
Hypoplasia of thymus |
0 |
2/9 |
- |
- |
Intestinal malrotation or ectopic spleen |
0 |
1/9 |
- |
+ |
Cleft palate |
0 |
1/9 |
- |
- |
Radio-ulnar synostosis |
0 |
1/9 |
- |
- |
Pyloric stenosis |
0 |
0 |
+ |
- |
Pharyngo/laryngomalacia |
0 |
0 |
- |
+ |
Cholestasis |
0 |
0 |
- |
+ |
Our patient has substantial gastrointestinal and pulmonary complications which are novel features of 14q aberrations.17 Her GI issues include gastrostomy tube dependence for feeding intolerance, duodenal malrotation and gastroesophageal reflux disease are more significant than in other patients with abnormalities involving 14q. She has many features which have not been previously reported in patients with either 14q deletion or duplication including neonatal cholestasis with bile duct obstruction and paucity of bile ducts laryngomalacia, severe obstructive sleep apnea, tracheomalacia, pharyngomalacia and congenital cystic adenomatoid malformations.
Subtle differences in genotype may explain differences in clinical features between our patient and the previous case: the 14q duplication and deletion present in our patient were delineated at molecular level while the previous case was characterized at chromosomal level.17 Although both rearrangements appear to involve similar breakpoints on 14q, the difference between the two cases at the molecular level may be considerable. Alternatively, the origin of the 14q segment differs between patients: in our patient the aberration is novo while the previous case was of paternal origin. Differences in clinical features may be due to age: our patient is younger, so features may not have yet developed.
Three mechanisms for forming an inverted duplication adjacent to a deletion have been proposed: the presence of paracentric inversion in one of the parents, low-copy repeats-mediated non-allelic homologous recombination (NAHR), and U-type exchange.24,25 A disomic region exists between an inverted duplication and a deletion that is formed through either a parental paracentric inversion or recombination between inverted low copy repeats (NAHR), but not in 14q rearrangement derived through the U-type exchange.25,26 High resolution microarray analysis of our patient revealed an unanalyzed region between the inverted duplication and the deletion smaller than 12 kb. Therefore, it is unlikely that a disomic region is present in the interval supporting that the 14q inverted duplication and deletion in our patient is most likely resulted from a U-type exchange following a double-strand break.
Our patient has significant gastrointestinal and pulmonary defects which have not been previously described with 14q aberrations and is the first case of 14q terminal deletion and distal duplication in tandem to be characterized by high resolution microarray.
None.
None.

©2016 Santoro, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.
 International Childhood Cancer Day is observed on 15 February 2026 to raise awareness about childhood cancers and to highlight the medical, developmental, and supportive care needs of affected children and their families. This day emphasizes the importance of early diagnosis, pediatric care, and continued research to improve survival and quality of life in children with cancer.
Researchers and healthcare professionals are encouraged to submit their original research articles, reviews, and clinical studies related to pediatric oncology, neonatal care, and child health. Manuscripts submitted on the occasion of International Childhood Cancer Day will be eligible for a special publication discount of 30–40% in the Journal of Pediatrics & Neonatal Care (JPNC).
.
International Childhood Cancer Day is observed on 15 February 2026 to raise awareness about childhood cancers and to highlight the medical, developmental, and supportive care needs of affected children and their families. This day emphasizes the importance of early diagnosis, pediatric care, and continued research to improve survival and quality of life in children with cancer.
Researchers and healthcare professionals are encouraged to submit their original research articles, reviews, and clinical studies related to pediatric oncology, neonatal care, and child health. Manuscripts submitted on the occasion of International Childhood Cancer Day will be eligible for a special publication discount of 30–40% in the Journal of Pediatrics & Neonatal Care (JPNC).
.