Journal of
eISSN: 2373-437X


Review Article Volume 3 Issue 2
1Department of Biological Sciences, Rani Durgawati University, India
2Department of Zoology & Biotechnology, Govt. Modal Science College, India
3Department of Zoology, Govt. O.F.K. College Khamaria, India
4Madhya Pradesh Council Sciences and Technology (MPCST), Govt. M.P. Bhopal, India
5Biodiversity Board, State Forest Research Institute (SFRI) Polypathar, India
Correspondence: Patel RN, Department of Biological Sciences, Rani Durgawati University, India, Tel 91-7694036235
Received: January 13, 2016 | Published: February 25, 2016
Citation: Patel RN, Patel DR, Bhandari R, Silawat N, Homkar U, Sharma J (2016) Upcoming Plant Pathological Techniques in the Disease Diagnosis. J Microbiol Exp 3(2): 00087. DOI: 10.15406/jmen.2016.03.00087
Serodiagnostics for the detection of plant pathogens particularly the plant viruses started in the fifties and had a quantum jump in application after the introduction of enzyme conjugated antibodies in enzyme-linked immunosorbent assays (EIA) in the 1970s. Improvements and modifications in the original EIA made its application routine in the large scale indexing of vegetatively cultivated crops. It is still the methods of choice for many purposes. However, its limited sensitivity as compared to nucleic acid based diagnostics particularly PCR restricts its use in critical decision making related to pathogens of quarantine significance like ring rot of potato caused by Corynebacterium sepedonicum, tuber necrosis strains of PVY (PVYNTN), etc. Serodiagnostics due to limited sensitivity and specificity in the detection of fungal and bacterial pathogens has not become popular for the detection of many of these pathogens. Recent developments in the field of genomics of microorganisms and PCR based assays of microbes have revolutionized diagnostics. A combination of immune capture of pathogen and PCR has made the detection simpler and more sensitive than PCR alone. Combination of immune capture with multiplex PCR further reduces cost of detection. Combination of nucleic acid probes and ELISA has the potential of its adoption for large scale indexing of propagating stocks. WTO agreements related to trade in agricultural commodities need internationally acceptable diagnostic tools for the detection of pathogens of quarantine significance. An internationally recognized monitoring organization is the urgent need. The organization will be responsible for the evaluation of reagents and protocols for diagnostics and their regular update. The issue was mooted in a recent symposium titled “Moving Plant Disease Detection from the Ivory Towers to the Real World” where a ring test process was described to validate and standardize new diagnostic procedures.1 As more and more techniques are validated by the scientific community and their reliability and cost-effectiveness demonstrated, molecular based techniques will play a greater and more important role in the detection of pathogens.
There have been phenomenal developments in the discipline of plant pathology aided primarily by the revolution in molecular biology, biotechnology, genomics and information technology. The more pronounced advancements in plant pathology have been in the field of diagnostics of pathogens. Initial push in diagnostics was provided by sero diagnosis in the sixties.
It was accelerated by enzyme immuno assay and further speeded up by nucleic acid hybridization and finally by polymerase chain reaction. Till almost mid 1980s, the main aid to confirm phytoplasmal infection was electron microscopy of thin sections of phloem of infected tissue. Similarly, detection of fungal and bacterial pathogens till 1970s was through biological, in vitro culture and microscopic techniques.
A successful strategy of disease management has to heavily rely on the identity of the pathogen and its characteristics viz. life cycle, mode of spread, weak links in the life cycle, alternate hosts, etc. Thus, identity of the pathogen is the first and most important step in disease management. It is particularly so in case of vegetatively propagated horticultural crops.
Developments in genetic engineering and resultant ease in the production of recombinant pathogens with greater virulence2 coupled with new and emerging diseases particularly the viral ones are adding further significance to the pathogen diagnostics. Increase in international travel and exchange of goods in trade can lead to intentional or inadvertent introduction of pathogens of quarantine significance. Highly sensitive, reliable, high throughput diagnostic techniques are required to deal with Sanitary and Phytosanitary (SPS) issues in international trade in agricultural commodities.
An ideal diagnostic test should fulfil the following criteria viz. high detectability i.e. should be able to detect a very low concentration of the pathogen, high specificity i.e. specific only to the target, high sensitivity i.e. signal to reactant ratio should be high (as in ELISA), high accuracy (conformity to reference), simplicity (easy handling with minimum steps), rapidity, stability of the reagents, amenable to automation and reasonably low cost. However, none of the diagnostic tests meets all the above criteria and a combination of two supplementary tests is often required for taking decisions of sensitive nature.
Various recent developments in the field of plant pathogen diagnostics have been reviewed by Chu et al..3 Clark;4 Dehne et al.5 Duncan & Torrance;6 Hampton et al.7 Henson & French;8 Khurana & Garg;9 Khurana;10 Miller & Martin;11 Seemuller et al.12 Torrance;13 Louws et al.14 Martin et al.15 Schaad16 and Schaad et al.17 An overall review of advancements taken place during the last about three decades and currently underway are covered in this chapter.
Fungi being having a more complex genome and structure as compared to plant viruses, was less amenable to detection through serological means. Surface components in fungal structures are more complex and selection of antibodies highly specific for detection at sub-species level is a very laborious exercise. Therefore, sero diagnosis in fungi was used more for disease diagnosis than the specific detection of a pathogen infection like the latent infection of late blight pathogen in potato tubers. Use of serological techniques in fungal pathogen diagnosis was reviewed by Miller & Martin.11 Some of the instances where serodiagnosis was employed were immunocytochemical detection of root fungi in Rhododendron using colloidal gold particles coated with antibodies raised against the fungal pathogen(s),18 and in the immunobinding dot-blot assay.19
Monoclonal antibodies able to detect all isolates of Sirococcus strobilinus only from spruce and pine,20 and those directed against zoospores and cysts of Phytophthora cinnamomi and showing specificity at isolate, species and family level were reported by Hardham et al.21
‘Dipsticks’ where primary antibodies are bound to a membrane attached to the end of sticks were used to detect fungal pathogens in turf grass.22 The ‘dipstick’ is incubated with the extract of soil or plant material and the enzyme-conjugated antibody. The ‘dipstick’ is then rinsed and transferred to substrate solution. After a short incubation period, the ‘dipstick’ is dried, and the intensity of color development is quantified using a handheld deflect meter.
Nucleic acid based diagnosis
Several sets of universal primers for amplification of fungal sequences published by White et al.23 provided a basis for the development of fungal sequence database. Molecular analyses techniques with resolution higher than the species level have resulted in rapid expansion of population genetics studies. Molecular analyses are used to identify species or particular genotypes within a population. The topic of molecular detection of fungi was reviewed recently by Palfrey24 and Schaad et al.17
Use of PCR in the detection of fungal pathogens has witnessed tremendous increase. It has allowed for the differentiation between closely related species of some pathogens which was otherwise difficult on the basis of subtle morphological characteristics or differences in host range. Thus, real time PCR using TaqManTM probes has been used to detect Phytophthora infestans in potato and P. citricola in citrus, Diaporthle phaseolorum and Phomopsis langicola in soybean seeds, Helminthosporium solani , Colletotrichum coccodes, and Rhizoctonia solani in soil and potato tubers and Aphanomyces euteiches in alfalfa.17 In addition, a real-time PCR assay using molecular beacons was developed for Rosellinia necatrix, white root rot of fruit trees, real-time PCR was used to detect Pyrenophora spp. in barley seeds, Blumeria graminis f. sp. tritici on wheat , rice blast fungus Magnaporthe grisea and Cladosporium sp., Ramularia sp. and Microsphaera alphitoides on oak.17 Several real-time PCR assays have been developed using TaqManTM probes for Tilletia indica, causal agent of Karnal bunt of wheat25 and Phakopsora pachyrhizi which causes rust on soybean.26
Some of the nucleic acid based detection methods for fungi are enumerated below with respect to Phytophthora species.
Chemotaxis of zoospores and antibodies: Dipsticks coated with chemotractants that are used directly with a species-specific immunoassay can both attract and detect cinnamomi27 and P. nicotianae.28 This elegant and fast procedure can be used to monitor these species in soil. Note that zoospore chemotaxis can be an easy way to concentrate the inoculum on a small surface before running a molecular assay.
DNA probes: Plasmid libraries can be made with random DNA fragments of fungal pathogens and individual fungal DNA fragments in the plasmids of bacterial colonies can be tested for their specificity as probes. Three random chromosomal DNA clones hybridized only to parasitica when tested against 10 other Phyhtophthora spp.29 and two other clones were species specific to P. citrophthrora.30 DNA hybridization was used to detect P. parasitica directly from leaf disk baits.31
DNA sequences have been generated for phylogenetic analyses of Phytophthora spp.32-34 It is possible to select a specific sequence directly from an alignment to produce oligo-nucleotide hybridization probes. Such probes for detection of P. cinnamomi, P. palmivora, P. megakarya and P. capsici were prepared from alignments of ITS sequences.35
RFLPs: RFLPs of genomic DNA coupled with random probes were used to differentiate formae speciales and even isolates of 36,37 RFLPs of mitochondrial DNA (MtDNA) were used to differentiate formae speciales of Fusarium oxysporum.38 Most RFLP work with fungi has concentrated on MtDNA which upon cutting with a single restriction enzyme (with a 6-base recognition sequence) yields only 10-20 fragments that give distinct bands on agarose gels.
PCR random sequences: After partial sequencing of plasmid clones that were developed as specific probes for parasitica29,31 and P. citrophthora,30 specific PCR primers were designed for these two spp. And used successfully on tomato stems.39 PCR primers were designed from the partial sequence of a tandem repeat satellite DNA and were sued to detect P. infestans in potato leaves or tuber slices two days after inoculation.40 PCR primers were designed from RAPD fragments amplified from these fungi in inoculated plants.41
PCR-rDNA internal transcribed spacer: The region of the Phytophthora genome coding for ribosomal DNA (rDNA) has been extensively sequenced for phylogenies, and within rDNA region, the Internal Transcribed Spacer (ITS) region has been used preferentially for studies targeting species differences.
A primer pair species-specific for P. citricola was designed within the ITS1 and -2 regions and was used to detect this fungus in inoculated plants.41 A primer pair from the ITS1 and -2 regions was designed to amplify both Peronospora sparsa and P. cactorum directly from Rubus arcticus.42 Differentiation between the two species was accomplished by using another primer pair specific only to P. sparsa.
PCR-other genes or spacers: Targeting new genes will become easier as more genes are studied for phylogenetic purposes43 and as more data become available through projects such as the Phytophthora Genome Initiative.44
Several challenges have to be overcome for successful development of real-time PCR assays for fungal plant pathogens. The assays must have absolute specificity and should not detect amplicons from other closely related species or formae specialis. Once PCR primers and probes have been developed, the assays must be optimized and rigorously tested against other closely related species to prevent false positives. In addition, assays for fungal pathogens must detect the target pathogen at an acceptable level of sensitivity. Furthermore, efficient DNA extraction protocols need to be developed from infected plant material, and in some instances, from isolated spores.
Few, if any, PCR assays are used to identify fungal pathogens in disease diagnostic laboratories due probably to lack of funds to purchase necessary equipment, lack of adequately trained personnel, or the lack of a system or mechanism to validate assays for general use.
Nested Multiplex PCR: For simultaneous detection of more than one pathogen, specific nested primers are used after a first round of PCR with specific ITS primers and the technique is called ‘Nested Multiplex PCR’. It was used to detect the cereal pathogens namely Fusarium culmorum, F.avenaceum, F. graminearum, F. poae, Rhizoctonia cerealis, etc.15
Analysis of highly conserved rRNA gene sequences during 1980s45-47 laid the basis of phylogeny of mollicutes. Comparison of 16S rDNA among members of the class Mollicutes indicated that the mollicutes arose from a gram-positive clostridiumlike bacterial ancestor.45 Phylogenetic investigations of MLOs began in the late 1980s. Lim & Sears48 compared 16S rDNA sequences from an MLO belonging to aster yellows group with Acholeplasma laidlawii and an animal mycoplasma. They discovered that the MLO was more closely related to Acholeplasma than to the animal mycoplasma. Subsequent studies analyzing several ribosomal proteins confirmed the close phylogenetic relationship between this MLO and Acholeplasma.49,50
Global phylogenetic analyses of 16S rRNA and rp gene operon sequences showed the phytoplasmas formed a large monophyletic clade within the expanded Anaeroplasma clade.51 In the past 5 years (1995-2000), 20 subclades were recognized.12 These comprehensive phylogenies have formed a basis for classification of this uncultured plant pathogen group. The phylogenetic subclades coincide with 16S rRNA phytoplasma groups identified by RFLP patterns, validating the classification schemes that are based on RFLP analysis of 16S rDNA sequences.52
The 16S rRNA sequence similarities among subgroup members of a given phytoplasma 16S rRNA group range from 95% to >99%, whereas the similarities range from 88 to 94% between two distinct 16S rRNA groups.52 However, there are examples of bacterial strains that are readily classified as distinct species by conventional approaches (e.g. DNA homology and phenotypic characters) but cannot be distinguished by analyses of their 16S rRNA sequences, because some of them share 99% or higher 16S rRNA sequence similarity.53
Additional phytogenetic markers such as rp genes, the tu f gene, 23S rRNA gene, and 16S/23S rRNA intergenic spacer sequences have been used as supplemental tools for differentiation of phytoplasmas.54
Issues related to diagnostics
As already stated under introduction, none of the diagnostic techniques meets all the requirements. Molecular technology based techniques particularly PCR and combination of PCR and serology have shown potential of fulfilling many of the requirements of an ideal diagnostic tool. Martin et al.15 have remarked “If the concept of a universal tool for pathogen detection is ever realized, it will very likely be a molecular diagnostic technique, or a combination of molecular techniques based on highly conserved genomic sequences”. Some of the important issues related to diagnostics are discussed below:
Cost
A diagnostic test besides being sensitive and reliable should also be economical particularly when a large number of samples have to be tested. Key elements of molecular diagnostic technologies are often patented and involve complex protocols requiring expensive reagents as in PCR. PCR assay can be somewhat simplified by combining immuno-capture (IC) of the target pathogen (particularly virus) with reverse transcriptase (RT)-PCR. IC-RT-PCR is not only simpler and less costly but also more sensitive.55 Simultaneous detection of several target pathogens through multiplex PCR is also cost saving. However, loss in sensitivity may occur when multiplex IC-RT-PCR is employed for detection of closely related viruses.56 It can be overcome by standardizing the detection protocols.
If target pathogen is any member of a particular virus group rather than a specific virus degenerate primers-based PCR can be employed advantageously. Thus, a single pair of degenerate primers targeted to conserved regions in heat shock protein (HSP) 70 gene of the Closterovirus has been used to detect several of its members.57 Similarly, conserved region extending from the coat protein gene to the NIb gene in Potyviridae58 and the conserved 3′ region of the S RNA of Tospovirus59 have been used to devise degenerate-primer based PCR detection of the members of these virus groups.
Sensitivity
Sensitivity of detection is of paramount importance for the testing of pathogens of quarantine significance. But, serology-based detection of some pathogens is limited to certain months of the year and to certain parts of the plant particularly in case of some woody plants. PCR detection is helpful in these situations since sensitivity of the test allows detection of the pathogen in most tissues.60 Reliability of detection is further enhanced by employing IC-RT-PCR with ELISA.55
Specificity
Detection of a specific strain of a particular microorganism or distinction of different strains is of major concern for pathogens of quarantine significance. Specific diagnostic tests are essential for bacteria such as Erwinia, which has many species and strains with varying host range and specificity. Digoxigenin-labeled DNA probes and ligase chain reaction (LCR) assays have been used for highly specific diagnosis of Erwinia.61,62 Highly specific primers are used to distinguish strains of PVY in PCR assays.
Validation of diagnostic test
Diagnostic reagents and protocols need validation for international acceptance. It is particularly the case for the testing of pathogens of quarantine significance. The evaluation process must be monitored by an internationally credited organization. Once standards are agreed upon, protocols must be revised regularly to avoid becoming obsolete. Free international trade without non-tariff barriers under WTO will be possible only when properly validated pathogen detection technologies are used.
Impact of diagnostics on epidemiology and disease management
Epidemiology: Epidemiology deals with the disease cycle where intimate interactions occur amongst the pathogen, host and environment. Diagnostics plays an invaluable role in the study of these interactions. Following aspects of epidemiological studies will be discussed.
Source of inoculums: Identification of the source of inoculums is very essential in the study of a disease cycle. Severe late blight epidemics occurred in Europe and North America during 1990s due to the appearance of A2 mating type. Global migration of Phytophthora infestans during 1980s was investigated employing molecular markers, which demonstrated that strains in the Pacific Northwest USA during the late 1980s were recent introductions from Northwest Mexico. These strains had a unique mitochondrial heliotype that was detected with a PCR-based technique.63
Understanding population genetics: Molecular markers are helping in accurate understanding of basic population genetics of several plant pathogens. Analysis regarding the origin of A2 mating type of infestans indicated it to be a recent introduction into Europe from Mexico and that the recent immigrant strains were more aggressive.64,65
Near real time detection: PCR-based assay due to very high sensitivity can detect arrival of a pathogen before the appearance of symptoms and the information can be used in disease forecasting schemes.
Environmental effects on pathogens and hosts: Whether the environment affects the host or the pathogen or both and how can be studied with micro arrays with genes of both the host and the pathogen.
Disease management
Diagnostics plays a very important role in disease management. Detection of the source of inoculum or the pathogen before appearance of disease symptoms helps in managing the disease through eradication of the source of inoculums or through forecasting of the disease. In vegetative propagated crops like potato and many other horticultural crops, selection of healthy planting stocks is very essential. All the programmes of supply of certified/healthy propagating stocks need sensitive and reliable diagnostic techniques to select disease-free material. Diagnostics plays a very important role in plant quarantine regulations meant for checking the entry of objectionable pathogens in an otherwise pathogen-free area.
Immunofluorescent assays have been employed to diagnose diseases caused by bacteria.66-68 Auto fluorescence of plant material and fungicides in soil samples, adsorption of tagged antibodies to soil particles, and potential cross-reactivity can affect interpretation of results and must be analyzed individually for each system.69
Monoclonal antibodies (MAbs) that were specific at the genus, pathovar and strain levels were produced against Xanthomonas campestris pv campestris (Xcc). An MAb able to distinguish Xcc isolates from non pathogenic xanthomonads, recovered from crucifer seeds, was reported by Alvarez et al.70 MAbs specific for Erwinia carotovora subsp. atroseptica (Eca) strains in serogroup I and XXII and non-reactive to E.carotovora subsp. carotovora (Ecc) were prepared by DeBoer & McNaughton.71 One MAb highly specific to Corynebacterium sepedonicum was used to detect latent ring-rot infection in potato by IFA staining.66
Nucleic acid based detection
Technological advances in molecular biology have radically changed our capacity to rapidly characterize and track microorganisms. Complete genomic DNA sequences of 18 microorganisms and an additional 56 under way72 have provided an extensive framework for a new era of microbial genetics, classification, and identification. These genomic approaches have also generated molecular tools for rapid and high resolution pathogen detection and disease diagnosis.
With the development of PCR, DNA-based techniques have rapidly become the preferred tool for identification of plant pathogenic bacteria. A large number of classical PCR primers for identification of important plant pathogenic bacteria are listed in the Laboratory Guide for Identification of Plant Pathogenic Bacteria73 and many more are becoming available. PCR-based assays offer many advantages over traditional isolation and immunological methods; most important are specificity and time.
Specificity of PCR depends upon the uniqueness of the sequences selected for primers and probes. Improvements in sequencing technologies are making the selection of reliable PCR primers routine.74
Real time PCR for detection of phytopathogenic bacteria is fast and may be very specific. But, it is often not very sensitive. Combination of PCR with isolation of bacteria on media whereby there is enrichment of the microbes greatly enhances their detection. This technique is termed BIO-PCR.75 In a BIO-PCR assay, infected/target plant extract is plated onto agar or added to liquid media and enriched for 15 to 72h. Since bacterial cells lyse during the initial denaturing step of the amplification, no DNA extraction is required.75 Some of the phytopathogenic bacteria for which PCR protocols have been developed include Pseudomonas syringae pv phaseolicola, Clavibacter michiganensis subsp. sepedonicus, Ralstonia solanacearum, R.solanacearum bv-2, Xanthomonas albilineans, Acidovorax avenae subsp. avenae, Agrobacterium tumefaciens and Escherichia coli.17 For ultra high sensitivity, BIO-PCR can be used with membranes.76 It employs surfactant-free membranes (Sartorius, Edgewood, NY, No.12587) made especially for BIO-PCR. The sample is filtered through the memberane to retain the target bacterium and the membrane is placed on semiselective agar media.After 1-3 days incubation, the filter is removed and placed into a microfuge tube with 50ul water and vortexed to suspend the bacteria. The sample is then used for direct PCR. As little as 1 to 3 cfu/ml of P. syringae pv. phaseolicola is readily detected.77 An added advantage of membrane PCR is that the sample is freed from PCR inhibitors and the main disadvantage is that the technique introduces a step for possible cross-contamination. An alternative to BIO-PCR for increasing sensitivity of PCR is to use immuno-capture PCR.78 Real time BIO-PCR protocols have detected 2cfu/ml of C. sepedonicus in potato tuber extract.79
In addition, multiplex PCR can be employed to detect more than one species of bacterium in the same reaction tube using probes labeled with different fluorescent dyes such as FAM, TET, TAM and ROX. To avoid cross-absorption, the wavelength of each dye must be well separated.
Specific recognition between the antibodies and their homologous antigens was recognized in the 50s. Various serodiagnostic techniques for virus detection are shown in Figure 1. Initially used serodiagnostic methods like double diffusion in gels, chloroplast agglutination, etc. were less sensitive and unsuitable for viruses like Potato leafroll virus which occur in a very low titre in infected tissue.
ELISA: A device to amplify the signal of antigen-antibody interaction for sensitive and reliable detection was needed and achieved by conjugating an enzyme with the antibody. The technique referred to as Enzyme-linked immunosorbent assay (ELISA) was published by Clark & Adams in 197780 for the detection of plant viruses. It is a solid phase procedure and is carried out in microtitre plates. In ELISA, enzyme conjugated antibody is made to bind its homologous antigen and the antigen-antibody complex is probed by adding enzyme substrate which upon enzymatic breakdown produces a coloured product (Figure 2A). Intensity of colour is the measure of relative concentration of the antigen and is in turn the function of antigen titre and time. Many formats of ELISA are in use and all are referred to as enzyme-immuno assays (EIA). Several modifications of the ELISA technique have been attempted for simplification and enhanced reliability. The use of fluorogenic substrates,81 modified antibodies F(ab)2-fragments,82 enzyme amplification83,84 or time-resolved fluorometry85 were some of the major modifications during 1980s. Indirect and direct antigen-binding dot enzyme immunoassays (Dot-EIA) on nitrocellulose or nylon membrane, in place of the polystyrene solid phase of ELISA, were useful further variants of the solid phase EIA.86,87
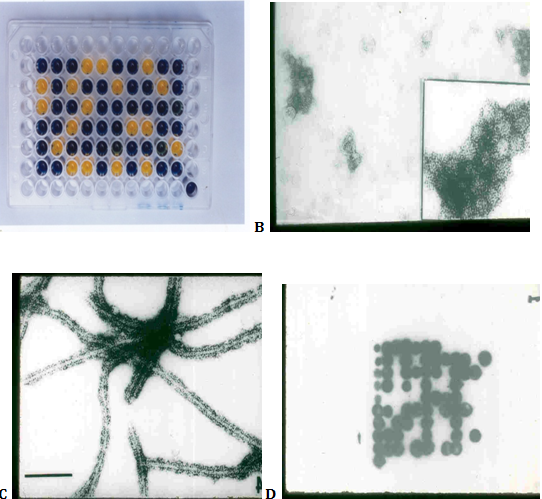
Figure 2 Microtitre plate showing dot-ELISA results of Potato virus X (PVX) detection by using penicillinase-conjugated antibodies; see loss of colour in positive samples (A); results of immuno electron microscopic virus detection showing clumps of Potato leafroll virus (PLRV) (B) and decorated virions of Potato virus Y (PVY) (C); results of nucleic acid hybridization technique for the detection of PVY where probe was labeled with 32P (D).
Various formats of EIA can be grouped under two categories of direct and indirect ELISA. It is direct ELISA when primary antibody-enzyme conjugate directly binds the antigen and is indirect ELISA if primary antibody-enzyme conjugate does not directly bind the antigen.
DAS-ELISA is more specific and is narrow spectrum i.e. may not detect distinct strains of the same virus while triple antibody sandwich (TAS-ELISA) is less specific, broad spectrum and more sensitive and helps in studying serological relationship among strains of the same virus or between different viruses.88
Reliability of ELISA results depends on antibodies of high affinity and specificity. Achievement of this objective has been attempted through the production of monoclonal antibodies (McAbs) of high affinity and specificity.
ELISA, due to its reasonably high sensitivity, reliability, specificity and amenability to automation has become the test of choice particularly for large-scale indexing of seed/propagative stocks like potato and many other horticultural crops.
Microarray-based multiplex ELISA is being adapted for disease diagnosis.89 It will modernize and enhance efficacy of hitherto used serological assays and specifically detect numerous antigens simultaneously.
Recombinant antibody ELISA (R-ELISA): This is the latest technique utilising recombinant antibody (Rab) produced in bacteria. RAbs are fragments of variable heavy and light chains of antibodies (scFv).Selected specific scFv fused with different recombinant-proteins, including alkaline phospotase (AP) with a high enzymic activity, can be used in ELISA e.g. detecting PLRV.
For viruses (black currant reversion associated virus) for which specific poly Abs could not be produced, scFv R-ELISA has proved effective. ScFv encoding DNA has indefinite storability. Production of scFv in E.coli takes 2-4 wks. MCAbs are expensive to produce and some hybridoma cell lines either die or lose antibody production during storage.
R-ELISA proved equally useful for diagnosis of Citrus tristeza virus (CTV) with numerous isolates differing biologically and serologically and genome sequence. Screening McAbs to CTV revealed that two McAbs (3DF 1 and 3 CA5) detect all CTV isolates. McAbs genes, subcloned from the hybridomas and expressed in E. coli as scFv fusion proteins and used in ELISA showed that it was as specific and sensitive as the ELISA with parental McAbs.90
Now Biochips (densely packed probe arrays) are used for pathogen diagnosis. They are very flexible and robust with potential of increased diagnostic output and reduced cost. Both nucleic acid based and antibody-based microarrays are being developed.
Immuno electron microscopy
First electron microscopic examination of antigen-antibody complex91 involved pelleting of virus-antibody clumps and resuspension of the pellet in neutral phosphotungstic acid before being applied to a support film (filmed EM grid) and dried. A positive result showed in the electron microscope as a clumping of the virus particles which were covered with antibodies. A quicker method was described by Ball & Brakke92 and was designated as leaf-dip serology. The method involved placement of a drop of diluted antiserum on a filmed grid and then dipping the freshly cut edge of a virus-infected leaf in the drop for a few seconds. The drop was then allowed to air-dry and then the grid was negatively stained. However, final image was having drawbacks due to drying of the virus-antibody mixture on the grid. Foundation of the present day IEM was laid by Derrick.93 The two remarkable improvements incorporated were coating of the filmed grid with antibodies which was then used to trap the virus and washing of the grid at two points-after antibody adsorption and after virus adsorption to the grid. Washing removed salts and other components and resulted in clear background (Figure 2B & 2C). Milne & Luisoni94 made further modifications to improve IEM. Immuno electron microscopy (IEM), if properly standardized may prove even more sensitive and reliable than ELISA.95,96 But, IEM has the drawbacks of heavy capital cost, low throughput and requirement of skilled manpower.
Nucleic acid based diagnosis
Nucleic acid based diagnostics may employ cloned nucleic acid probes which are complimentary to a specific segment of the viral genome or may employ amplification of a specific segment of the viral genome with polymerase chain reaction (PCR). PCR due to its phenomenal ability to amplify a given segment of DNA in a short time has revolutionized molecular biology and diagnostics.97
Polymerase chain reaction
Genome sequences are known for many plant viruses. Consequently primers specific for virus group and type are available for PCR diagnosis. Since majority of plant viruses possess plus-sense RNA genome, RT-PCR is used to detect/identify them. RT-PCR is not suitable for routine testing due to time-consuming gel electrophoresis prior to visualization.
Sufficient sequence data for diagnostic primer and/or primer/probe sets are available for thousands of plant viruses. Conserved genomic sequences are used to detect viruses at species level while the variable sequences are used to distinguish between viral strains. Generalized primers have been used to detect members of tymoviruses98 and Potyviridae.58
Real-Time PCR: As the name suggests, real-time PCR starts giving the result, if positive, as soon as the amplification of the targeted sequence starts. It avoids the running of amplified product on gel. It can use TaqManTM probes, fluorescent resonance energy transfer (FRET) probes, or molecular beacons to detect the production of amplicons. These methods are based upon the hybridization of fluorescently labeled oligonucleotide probe sequences to a specific region within the target amplicon that is amplified using traditional forward and reverse PCR primers. In TaqManTM system, an oligonucleotide probe sequence of approximately 25-30 nucleotides is labeled with a fluorochrome at the 5′end99 and a quencher fluorochrome at the 3′ end.35 The TaqManTM probe is degraded by the 5′ to 3′ exonuclease activity of the Taq polymerase as it extends the primer during each PCR amplification cycle and the fluorescent chromophore is released. The amount of fluorescence is measured/monitored during each amplification and is proportional to the amount of PCR product generated.
FRET probes require labeling of two adjacent oligonucleotide probe sequences within the PCR target fragment.100,101 Probe 1 contains a fluorescein label at its 3′ end whereas the second probe is labeled at its 5′ end with another label such as Light Cycler Red 640. The two probes must be designed so that when they hybridize to the amplified PCR product they are aligned head to tail to bring the two fluorescent dyes in close proximity to each other.101,102 The fluorescein dye attached to the first probe is excited by the light source of the appropriate wavelength, and it emits a green fluorescent light at a slightly longer wavelength. When the 2nd probe is in close proximity, the energy emitted by the first probe excites the Light Cycler Red 640 dye attached to the 2nd probe, and the red fluorescent light at longer wavelength is now emitted that can be detected at 640nm. Fluorescence is measured during the annealing step of each of the amplification cycles when both probes hybridize to the PCR amplicon.100,101
Real-time-RT- PCR assays have been developed for detection of a large number of plant viruses.103-106 These techniques are capable of detecting TSWV in single vector thrips107 and Plum pox potyvirus (PPV) in the aphid vectors.
Multiplex PCR is used to detect several viruses in a single reaction.59,108-111 Array or biochip technology which uses expression of multiple genes simultaneously can be developed for simultaneous detection of several plant viruses.
DIAPOPS (detecting immobilized, amplified products in a one-phase system)
It was used for the detection of PVY.112 In this, NucleoLinkTM strips (NUNC A/S, Raskilde, Denmark) are covalently coated with an oligonucleotide which serves as one of the two primers in the following PCR. During PCR, amplicons are thus covalently attached to the micro well surface. These amplicons serve as targets for a biotin-labelled probe subsequently detected in ELISA reader after adding suitable streptavidin-enzyme/substrate combination. For plant virus detection, an RT step prior to DIAPOPS is included.
Complementary nucleic acid probes
Nucleic acid probes are stretches of nucleotides complimentary to a specific region or whole genome of a virus or viroid. Thus use of nucleic acid probe entails molecular hybridization i.e. binding of two strands of nucleic acids (DNA, DNA and RNA, or RNA). This binding can be affected in solution (solution hybridization) or on solid support like filter paper or membrane (filter hybridization). Solid support hybridization is also called nucleic acid spot hybridization or NASH (Figure 2D).
Nucleic acid probes have been used for virus and viroid detection where large scale indexing of samples is required.113-115. Both radio-labeled and non-radio labeled probes (cDNA or cRNA) have been developed for the purpose.114,116 Sensitivity of detection of PVY was enhanced by increasing the size of the cDNA probe from ca 0.56 kb to ca 3.25 kb.117
Molecular beacons: They are hairpin shaped fluorescent oligonucleotide probes where the loop portion of the probe contains nucleotide sequences complimentary to the target amplicon. A fluorochrome is attached at the 5′ end of the probe and a quencher molecule is attached at the 3′ Due to complimentarity of nucleotides in the stem region of the probe, fluorochrome and quencher lie close to each other and as a result there is no fluorescence. However, when the probe hybridizes to the target amplicon during PCR amplification, the fluorochrome gets separated from the quencher and fluorescence becomes detectable.100,101

©2016 Patel, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.