Journal of
eISSN: 2373-437X


Research Article Volume 10 Issue 4
1Assiut University Mycological Centre (AUMC), Assiut University, Egypt
2Department of Microbiology, Faculty of Science, Taiz University, Taiz, Yemen
Correspondence: Osama Abdel-Hafeez Al-Bedak, Assiut University Mycological Centre (AUMC), Assiut University, Assiut 71511, Egypt, Tel +21007786262
Received: August 27, 2022 | Published: September 7, 2022
Citation: Al-Bedak OA, Sakr RS, AL-Kolaibe AMG. The microbial amylases: an overview with practical consequences and applications. J Microbiol Exp. 2022;10(4):130-134. DOI: 10.15406/jmen.2022.10.00363
The amylase enzymes work to convert molecules of starch or glycogen into molecules of glucose or maltose. Amylases are produced by a large number of living organisms, including bacteria, plants, and people. Here, we provide an overview of these essential enzymes, outlining their classification, mechanism of action, applications in industry, and production technologies such submerged fermentation (SmF) and solid-state fermentation (SSF). The formulas used to determine amylase activity are also emphasized in this essay. Every data item in this document is explained with an illustration. This technical study might be essential for the work of the amylase enzyme.
Amylases are enzymes that convert starch or glycogen into glucose or maltose units. Amylases are produced by a large variety of living organisms, including bacteria, plants, and humans. Bacteria and fungi generate amylase from the outside of their cells for extracellular digestion. When the insoluble starch is broken down, soluble byproducts like glucose or maltose are absorbed in their cells. According to the kinds of molecules that amylases transform molecules produced from broken-down starch into:
(1) α-amylase (alpha-amylase; EC 3.2.1.1)
Other names: alpha-amylase; glycogenase; α-amylase; endoamylase; Taka-amylase A; 1, 4-α-D-glucan
glucanohydrolase.
Reaction: Endohydrolysis of (1-4)-α-D-glucosidic linkages in polysaccharides containing three or more (1-4)-α-linked D-glucose units.
Note: reduces the viscosity of starch by randomly rupturing the bonds, resulting in a variety of sized chains of glucose. Acts in a random way on starch, glycogen, and associated poly- and oligosaccharides; reducing groups are freed in the alpha-configuration. The initial anomeric structure of the liberated free sugar group is what is meant by the term "alpha," not the linkage's hydrolyzed state (Figure 2).
(2) ß-amylase (Beta-amylase; EC 3.2.1.2)
Other names: beta-amylase; saccharogen amylase; glycogenase; beta amylase; 1,4- α-D-glucan maltohydrolase.
Reaction: Hydrolysis of (1-4)- α-D-glucosidic linkages in polysaccharides so as to remove successive maltose units from the non-reducing ends of the chains.
Note: Breaks the glucose-glucose bonds down by removing two glucose units at a time, thereby producing maltose. Acts on starch, glycogen-related polysaccharides and oligosaccharides to produce beta-maltose by inversion (Figure 1).
(3) Amyloglucosidase (AMG; EC 3.2.1.3)
Other names: glucan 1,4-α-glucosidase; glucoamylase; amyloglucosidase; gamma-amylase; lysosomal α-glucosidase; acid maltase; exo-1,4- α-glucosidase; glucose amylase; gamma-1,4-glucan glucohydrolase; acid maltase; 1,4- α-D-glucan glucohydrolase.
Reaction: Hydrolysis of terminal (1-4)-linked α-D-glucose residues successively from non-reducing ends of the chains with release of β-D-glucose (Figure 1).
Note: Most forms of the enzyme can rapidly hydrolyze 1,6- α-D-glucosidic bonds when the next bond in the sequence is 1,4, however, some types of amylases can hydrolyze 1,6- and 1,3-α-D-glucosidic bonds. This entry covers all such enzymes acting on polysaccharides more rapidly than on oligosaccharides. Breaks successive bonds from the non-reducing end of the straight chain, producing glucose.
Alpha-glucosidase (EC 3.2.1.20)
Other names: α-glucosidase; maltase; glucoinvertase; glucosidosucrase; maltase-glucoamylase; α-glucopyranosidase; glucosidoinvertase; α-D-glucosidase; α-glucoside hydrolase; α-1,4-glucosidase.
Reaction: α-Glucosidase catalyzes the hydrolysis of terminal 1, 4-linked α-D-glucose residues successively from the non-reducing ends of maltooligosacharides and to a lesser extent polysacharides with release of β-D-glucose. Most forms of the enzyme can slowly hydrolyze 1, 6-α-D-glucosidic bonds.
Qualitative detection of amylase activity
Suggested media can be used: a basal medium supplemented with 1 % soluble starch as sole carbon source is a good choice for either amylase activity or production. The following two suggested media can be used sufficiently for qualitative detection of amylase activity or used as a fermentation media for amylase production in submerged fermentation or solid-state fermentation.
Medium composition (g/L): Soluble starch, 10; NaNO3, 2.0; KCl, 0.5; MgSO4 .7H2O, 0.5; K2HPO4, 1.0;
FeSO4 .7H2O, 0.01; agar, 15.0.
Medium composition (g/L): Soluble starch, 5.0; peptone, 5.0; yeast extract, 5.0; MgSO4.7H2O, 0.5;
FeSO4.7H2O, 0.01; NaCl, 0.01 and agar, 15.0.
Reagent: 0.25 % aqueous iodine solution: iodine, 0.25; potassium iodide, 0.75 and dist. water, 100 ml.
Procedure 1: Petri dishes containing the medium are inoculated with the tested microorganisms and then incubated at 35 °C for 72 h in case of bacteria or incubated at 30 °C for 7 days in case of fungi.
Procedure 2: A 50-100 µl spore suspension (prepared in 10 % tween 80) are inoculated in wells (5 mm in diameter) on the agar medium and the plates are incubated for 48 hours at 30 °C. After incubation period the plates are flooded with the iodine solution.
Result: Clear zone around fungal or bacterial colonies against blue color of the medium indicates amylase activity (Figure 2).
The same medium that was used to check for amylase activity may be used to manufacture amylase. One mL of spore suspension containing 1 × 106 spore/ml of 48-day-old bacterial cultures or 7-day-old fungal cultures is planted into each Erlenmeyer conical flask containing 50 mL of the fermentation medium. The flasks are incubated under shacking conditions for 3-5 days for bacteria at 37 °C and 120-150 rpm, or for 7-10 days for fungi at 30 °C.
The above-mentioned fermentation medium can be utilized to wet the solid substrate used for amylase synthesis. Fermentation can be carried out in Erlenmeyer conical flasks or fermentation flasks. The fermentable solid substrate is put in the flasks in a 1:50 or 1:25 ratio (250-mL conical flask contains 5 or 10 g of the solid substrate). The moisture level should be regulated to 70-80%. Following inoculation with the examined organism (fungi or bacteria), the flasks are incubated under static conditions at the same temperature as in SmF.
The cell-free supernatant is obtained in the case of submerged fermentation by centrifugation at 10,000 rpm for 10 min at 4 °C. While, in SSF, a buffer solution is used to isolate the enzyme from the fermented slurry before it is finished to a predetermined volume. The clear supernatant, which is used as a source of amylase in the assessment of amylase activity, is then obtained by centrifuging the enzyme solution at 10,000 rpm for 10 minutes.
Amylase assay can be carried out according to the method described by.13 The reaction mixture composed of 0.9 ml of 1 % starch solution (in 50 mM citrate buffer; pH 5) and 0.1 ml of cell-free supernatant (or 0.5 ml enzyme + 0.5 ml substrate). The reaction is completed at 50 °C for 10 min.5 This is followed by the addition of 2 ml of 3, 5-dinitrosalicylic acid (DNS) to the contents to eliminate the reaction, and then boiled in water bath for 10 min and cooled at room temperature (Dinitrobenzoic acid can be used as an alternative of Dinitrosalicylic acid and it gives the same results as well). Absorbance of the color developed is measured at 540 nm. The amount of reducing sugars liberated is quantified using standard curve of glucose (Figure 3).
Dinitrosalicylic acid (DNS) Reagent
|
Distilled Water |
1416 ml |
|
3,5-Dinitrosalicylic acid |
10.6 g |
|
NaOH |
19.8 g |
|
Dissolve above, then add |
|
|
Rochelle salts (Na-K tartarate) |
306 g |
|
Phenol (melt at 50 °C) |
7.6 ml |
|
Na metabisulfite |
8.3 g |
Glucose calibration curve (Figure 4)
0.1 g glucose is dissolved in 100 ml DI water as stock solution
|
Glucose (ml) |
DI Water (ml) |
DNS (ml) |
DI Water (ml) |
Absorbance Glucose (540 nm) |
|
0 |
1 |
2 |
7 |
0 |
|
0.2 |
0.8 |
2 |
7 |
0.127 |
|
0.4 |
0.6 |
2 |
7 |
0.418 |
|
0.6 |
0.4 |
2 |
7 |
0.536 |
|
0.8 |
0.2 |
2 |
7 |
0.737 |
|
1 |
0 |
2 |
7 |
0.988 |
One unit of amylase activity is defined as the amount of enzyme that liberate 1 µmole of glucose equivalent per minute under the standard assay conditions.8 Enzyme production and activity can be calculated as the following equations.1,2
Glucose concentration =
Amylase concentration =
If the substrate is used at 1 % concentration, then the enzyme concentration can be calculated per gram dry substrate (gds) as the following equation:
Amylase concentration =
Amylase activity =
Amylase specific activity =
Where: DF = the dilution factor for enzyme; x = the volume of enzyme used; y = the volume of hydrolysate used for assay of reducing sugars; t = the time of hydrolysis; slope is determined from a standard curve (= 1.0472).
1- Manufacture of high fructose containing syrup and maltose
High fructose containing syrups (HFCS) 42 F (Fructose content equal to 42%) is prepared by enzymic isomerization of glucose with glucose isomerase (Figure 5). The starch is first converted to glucose by enzyme liquefaction and saccharification.12,17
2- Manufacture of oligosaccharides mixture
Oligosaccharides mixture (Maltooligomer mixture) is generated by digestion of corn starch with α-amylase and β-amylase (Figure 6). The maltooligomer mix is a brand-new item on the market. Typically, it is composed of the following: maltotetrose and bigger malto-oligosaccharides, 14%; maltotriose, 37.5%; maltotriose, 46.4%; and glucose, 2.2%. 20
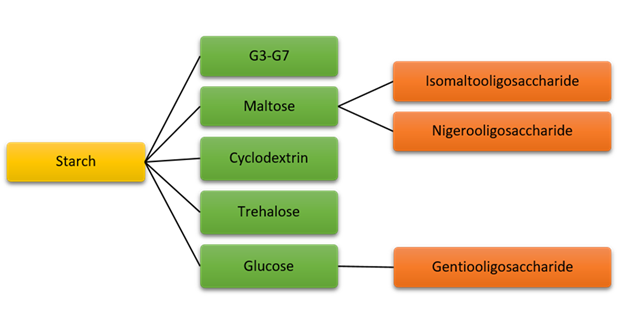
Figure 6 Microbial enzymes for the production of Starch related oligosaccharides (1) Malto-oligosacccharides forming amylase, (2) α-Glucosidase, (3) cyclodextrin glycosyltransferase (CGTase), (4) maltooligosyltrehalose synthase (MTSase) & maltooligosyltrehalose trehalohydrolase (MTHase) (5) ß-Glucosidase.
3- Bread and chapatti industry
Amylases may break down starch and produce microscopic dextrins that can be utilized by yeast. Wheat flour contains alpha-amylases, which break down broken starch into tiny dextrins, enabling yeast to function continuously during dough fermentation, proving, and the initial stages of baking. The consequence is an improvement in the bread's volume and crumb quality (Figure 7A). Additionally, the small oligosaccharides and sugars produced by these enzymes, such glucose and maltose, encourage the Maillard reactions that result in crust browning and a delicious baked flavor.18
4- Chocolate industry
Amylases are added to cocoa slurries to create chocolate syrup (Figure 7B), which does not thicken since the chocolate starch is dextrinizing.6
1- Removal of starch sizer (Desizing) from textiles
In textile weaving, starch paste is used to warp the fabric (Figure 8). While weaving, this strengthens the cloths. Additionally, by giving the surface of the string some softness as a result of the lay down wrap, it prevents string loss due to friction, cutting, and the development of static charge on the string. Following weaving, the cloth is starch-free, scoured, and coloured. Alpha amylase is typically employed to remove starch from textiles.3,9,18
2- Liquefaction
In order to achieve liquefaction, insoluble starch granules are dissolved in aqueous solution and then partially hydrolyzed using thermostable amylases (Figure 9). The starch suspension for liquefaction in industrial processes is more than 35%. The result is an extremely high viscosity after gelatinization. When thermostable -amylase is used as a thinning agent, the viscosity is reduced and some of the starch is partially hydrolyzed. As a result, further freezing avoids starch retrogradation.14,18,21
1- Direct starch fermentation to ethanol
Amylases are used in both the brewing and alcohol industries. Such systems have the advantages of homogeneous enzyme activity in mashes, speeding up saccharification, alcohol production, and yeast growth.7,19 Sugarcane, sugar beets, and sweet sorghum, which contain significant levels of sucrose, can be used bioethanol production. Then, an aqueous raw sugar juice (sucrose syrup) is extracted using the conditioned sugar solution (Figure 10). Enzymatic liquefaction and saccharification are needed to produce glucose syrup from starch crops like maize or wheat.18,22
2- Bio-fuel production
In the long term, fossil fuels cause greater harm to the ecosystem through pollution. Recent decades have seen a significant increase in interest in biofuels as a result of rising gasoline costs and environmental concerns. The most popular kind of biofuel is ethanol fuel. Renewable resources may be used to make ethanol, including agricultural crop byproducts and garbage. Alpha amylase, glucoamylase, cellulase, and other enzymes are required for the formation of fermentable sugars, which are subsequently transformed into ethanol.11,18
Alpha amylases are employed to help with its removal since starch paste, when used as a mounting adhesive and modified with additives like protein glue or alum, typically causes embrittlement in paper. The raw starch used for sizing and coating the paper was digested by alpha amylase instead of expensive chemically altered starches. Because of this, starch is frequently used in many paper-sized press publications.15,18
The detergent business depends on the enzyme -amylase. It is frequently used for bleaching without colour degradation and to boost the detergency of laundry bleach formulations. In the formulation of laundry detergent bars, the enzyme's presence stabilizes the bleach agent and retains the bleach's effectiveness. The feed has a lot of starches or very little else. Alpha amylase can thus be added to the food to increase its nutritional value.4,16,18
None.
Author declares that there is no conflict of interest.

©2022 Al-Bedak, et al. This is an open access article distributed under the terms of the, which permits unrestricted use, distribution, and build upon your work non-commercially.